
Sachiko Sato
Research Centre for Infectious Diseases, Faculty of Medicine, Laval University, Quebec City, Canada. Ph.D, Pharmaceutical science
Sachiko Sato graduated from Faculty of Pharmaceutical Science, Chiba University. She joined as postgraduate student in the laboratory of Dr. Akira Kobata, the Institute of Medical Science, the University of Tokyo, Japan in 1987. She also worked in the laboratory of Dr. R. Colin Hughes, MRC: National Institute for Medical Research in London, UK, where she first encountered a cytosolic mammalian lectin, now called galectin-3. She obtained her Ph. D. from the University of Tokyo in 1994. As postdoctoral fellow in the laboratory of Dr. Ron Kopito, Stanford University, she was involved in the work on cystic fibrosis. She became principal investigator of the laboratory of glycobiology in Research Center for Infectious Diseases, and assistant professor of the Faculty of Medicine, Laval University, Quebec, Canada in 1999 and is full professor since 2010. She is also director of the Bioimaging platform since 2003.
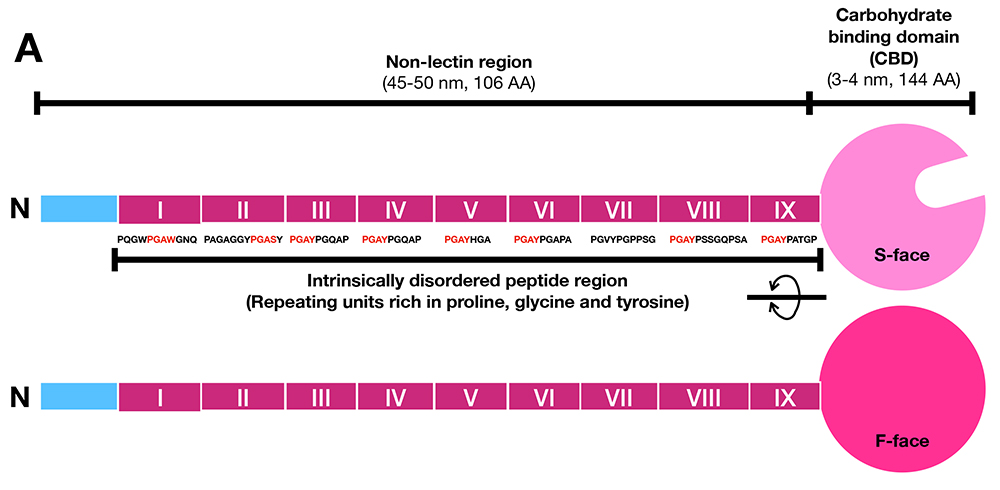
Galectin-3 is one of the most extensively studied members of the galectin family along with galectin-1. Galectin-3 is involved in a wide variety of functions and places, in vitro in both intracellular and extracellular spaces and in vivo in various organs. As mentioned in Part 1, galectin-3 contains an “intrinsically disordered peptide region”(Note 1) with repeated peptide sequences, and the total length of these peptides is almost the same as the carbohydrate-binding domain (CBD) at the C-terminus (Figure 3) 1,2. Notably, this region is not found in other animal lectins. Most of the reported activities of galectin-3 require this intrinsically disordered region. Galectin-3, which usually exists as a monomer, oligomerizes in a manner that is dependent on this region after binding to its glycan ligand. Therefore, the activity of galectin-3 is thought to be regulated primarily by intrinsically disordered region-dependent oligomerization. In Part 2, I would first like to delve deeper into the mechanism by which the unique intrinsically disordered region is involved in the oligomerization of galectin-3 by referring to the latest reports. Then, I will explore the relationship between the oligomerization of galectin-3 and its diverse functions.
Note 1: The galectin-3-specific N-terminal region is usually called the N-terminal nonlectin domain; however, the term “domain” should only be used for polypeptides that have formed secondary or tertiary structures. Thus, similar to Part 1, I will use the term “region” instead of domain. In addition, I refer to the repeated region within the N-terminal nonlectin region as the intrinsically disordered region.
Notable is the increasing number of reports suggesting that glycan-bound galectin-3 undergoes liquid–liquid phase separation (LLPS) through oligomerization in a manner dependent on the intrinsically disordered region at the N-terminus3–7. In Part 1, LLPS was described as a membraneless organelle. The space where LLPS occurs is a reversible condensed space where various biological reactions can efficiently occur or be controlled. The involvement of galectin-3 in the formation of this space is likely to be closely related to the following points: (1) galectin-3 has a wide variety of activities, (2) galectin-3 lattice formed on the cell membrane dynamically regulates various biological reactions (such as signal transduction and clathrin-independent endocytosis), and (3) most of the galectin-3 ligands required to induce the activity are not identified (because they cannot be narrowed down to one, they are ambiguous and difficult to clarify in detail).
In Part 2 of the forum, I will first discuss the similality between the LLPS already established in other systems and the behavior of galectin-3 bound to the cell surface. Next I will introduce recent papers on galectin-3 and LLPS and take advantage of the forum’s characteritics to further examine and explore on the activities of galectin-3 from the perspective of LLPS. There is a report suggesting that the CBD of galectin-3 interacts with the CBD of other galectins. Thus, I would also like to ponder the potential of a galectin network being involved in galectin-3-initiated LLPS, hoping that such a possibility will be investigated or explored in the future.
Similar to Part 1, as I have referred to papers centered around LLPS in writing this forum, I would like to mention in advance that I have not been able to cover all papers related to the oligomerization of galectin-3, which has a history of over 30 years.
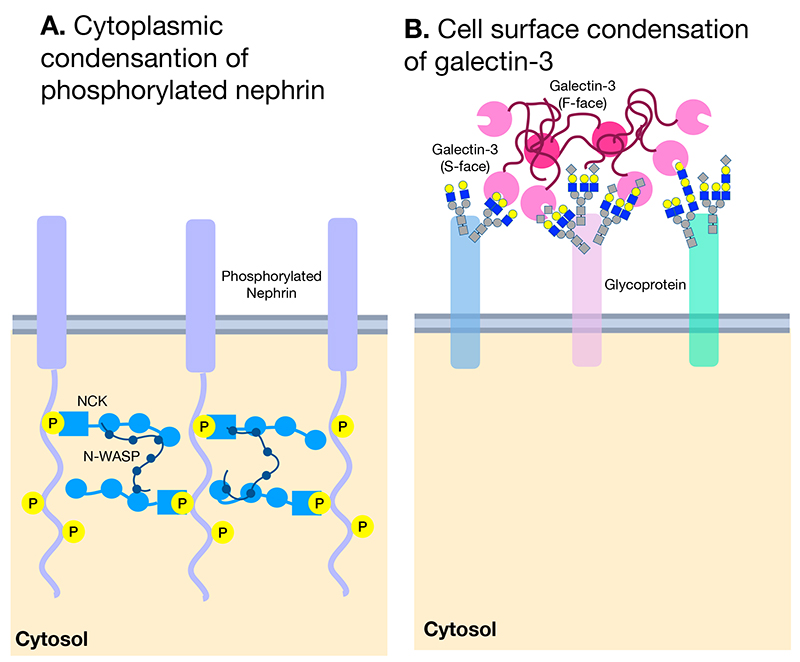
Inside cells, there are spaces such as stress granules and nucleoli where RNA and proteins are condensed despite the absence of a membrane. Recent studies have revealed that these are formed by LLPS occurring in three-dimensional (3-D) space. On the other hand, it was not clear until recently whether LLPS occurs in a 2-D space such as the cell membrane(Note 2). In 2014, Michael Rosen’s group demonstrated the occursence of LLPS on 2-D membrane by using phosphorylated nephrin and its cytoplasmic binding partners Nck and N-WASP, which carry an intrinsically disordered peptide region, on artificial membranes (Figure 2A) (8). Furthermore, the report suggests that in the presence of the Arp2/3 complex, this trimolecular clustering promotes the assembly of local actin filaments on membranes.
Note 2: In a 2-D space, such as on the cell membrane, membrane proteins have dynamic movement only in the XY direction since part of the proteins are within the lipid bilayer. This means that their movement is limited to diffusion in the 2-D direction only. On the other hand, in the case of soluble proteins, their dynamic movement is in the XYZ direction, which means they can diffuse in 3-D space.
In considering the relationship between galectin-3 and LLPS, there are several important points to note in this report. First, LLPS occurs suddenly when the concentration of the above three molecules reaches a certain value. Second, the propensity for LLPS to occur depends on the fact that all three molecules involved in the process exhibit multivalency in binding. Nephrin is phosphorylated at multiple sites, Nck possesses multiple binding sites to both phosphorylated nephrin and N-WASP, and the binding of proline-rich N-WASP is also multivalent (Figure 4A)8. Third, this trimolecular LLPS cluster is highly mobile, with the molecules inside undergoing exchanges between the cluster and its surroundings. Forth, most importantly, the concentration of the three molecules required for LLPS is 30 times lower in a 2-D environment (i.e., on the membrane) compared with a 3-D environment5,8,9.
In 2015, Dennis drew attention to these characteristics, proposing that the galectin lattice10 they had suggested since 2000 is the LLPS occurring on the extracellular side of the plasma membrane3. The illustrations in Figure 4A and 4B compare the similarities between the LLPS on the cytoplasmic side of the plasma membrane suggested by Rosen’s study and the galectin lattice on the extracellular side suggested by Dennis. Previous works by Dennis’ group had already suggested that the galectin lattice slows the lateral movement of its bound membrane proteins, while they are also mobile11,12 and undergo exchanges between the clusters and the surroundings13. Furthermore, since galectin-3 molecules oligomerize after binding to cell membrane glycoproteins, the binding is multivalent, and the region of galectin-3 required for this oligomerization is the intrinsically disordered region, which is proline-rich. The features suggested by Rosen8, where a phosphorylated membrane protein involves in LLPS together with a soluble binding partner and a protein with an intrinsically disordered region, are highly similar to ones of galectin-3 lattice where soluble galectin-3 binds to glycans of membrane glycoproteins at multiple sites through oligomerization in a manner dependent on the proline-rich intrinsically disordered region.
In 2017, Huang’s group in Taiwan performed further NMR structural analysis of galectin-3 at two concentrations (40 µM and 400 µM)4. It was reconfirmed that galectin-3 exists as a monomer at 40 µM but as an oligomer at 400 µM in the absence of sugar ligands. It was also confirmed that the N-terminal intrinsically disordered region alone oligomerizes whereas CBD remains as a monomer. In Part 1, I introduced a paper by Hughes' group14, which took images of galectin-3 and the N-terminal intrinsically disordered region using electron microscopy. Whereas galectin-3 does not form oligomers unless it binds to glycan; the images showed that the N-terminal intrinsically disordered region alone was able to form oligomers. Based on this, they speculated that the CBD that is linked to the N-terminal intrinsically disordered region may inhibit its tendency to undergo oligomerization. The result of Huang's group is consistent with the discussion in this paper14. At 40 µM, similar to previous studies, the molecular size of galectin-3 is smaller than expected. The intrinsically disordered region repeatedly binds and dissociates with the region of amino acids 200–220 of the F-face [the opposite face of the sugar-binding face (S-face)] at a high rate. The strengh of this interaction increases as the number of repeat structures in this region goes up. The same characteristic was also observed at 400 µM, where oligomerization occurs, suggesting that in the oligomers, several intrinsically disordered regions associate with and dissociate from one CBD at a high rate. In other words, the interaction seen here differs from the one-to-one, key-and-keyhole specific binding where a specific region binds to a specific region. Instead, several intrinsically disordered regions interact at a high rate with the specific site (amino acids 200–220 in the F-face of CBD) in a one-to-many manner as if they are scanning. Such dynamic interactions (sometimes called fuzzy interactions) are common in interactions within the LLPS involving intrinsically disordered regions15,16. Indeed, Huang et al. showed for the first time that the intrinsically disordered region induced reversible LLPS in a temperature-dependent manner.
These results indicate that the physical properties of galectin-3 have the potential to induce LLPS. However, the conditions in this report used a very high concentration of galectin-3 in solution (the 3-D) without its glycan ligands, therefore, differ considerably from the physiological conditions under which galectin-3 functions; the conditions in which it binds to membrane glycoproteins in 2-D space at concentrations below a few µM. However, as suggested by Rosen5, if the protein concentration that causes LLPS in 2-D space is more than 30-fold lower than in 3-D space, the concentration that can induce LLPS drops to around 10 µM in 2-D space. Furthermore, galectin-3 binds to several glycoproteins on the membrane, likely resulting in 10 µM or more local concentrations. Because the LLPS occurs rapidly when the required protein concentrations reach certain critical concentrations8, it is very likely that galectin-3 also induces LLPS on the membrane under physiological conditions. Let us look at two more recent papers exploring this possibility further.
In 2020, Huang‘s group showed that at 40 µM, galectin-3 but not the CBD induces the LLPS in the presence of micelles of lipopolysaccharide (LPS)6. This LLPS is reversible since it disappears in the presence of lactose. Galectin-3, in which all Ys and Ws in the PGxY(Note 3) or PGxW repeat in the intrinsically disordered region are mutated to Gs, neither induces LLPS nor interacts with the F-face of CBD. Therefore, it is suggested that the aromatic residues within the intrinsically disordered region are involved in the induction of LLPS. They point out that the theoretical galectin-3 concentration bound on the LPS micelle surface is as high as 15 mM when the molecular size of galectin-3 is considered. That is, galectin-3 binding to glycans on the membrane could easily reach the galectin-3 critical concentration at which LLPS could be induced.
Note 3: Proline (P), glycine (G), tyrosine (Y), and tryptophan (W)
In 2021, Tai and Zhou’s group investigated whether the interaction of galectin-3 with membrane glycoproteins CD7, CD45, CD71, and CD146(Note 4) induces LLPS in solution7. Galectin-3 induced LLPS in a concentration-dependent manner, although the degree of induction differed slightly depending on the type of glycoprotein. LLPS was disrupted by the addition of a galectin-3-binding antagonist, lactose. Galectin-3 mutant protein (R186S) with weak glycan-binding activity and partially truncated galectin-3 with a shortened intrinsically disordered region (six out of nine repeat structures are deleted) did not cause the LLPS. In addition, galectin-1 and -2 did not induce LLPS under the same conditions. These results suggest that after binding to glycans, galectin-3 induces LLPS in a manner dependent on the intrinsically disordered region. Most notably, LLPS does not occur with the intrinsically disordered region or CBD alone in the presence of those glycoproteins; however, when the intrinsically disordered region and CBD coexist, LLPS occurs. This suggests that LLPS can be induced even if these two regions are not necessarily connected as one molecule. On the other hand, CBD with a mutation (L203A) that cannot interact with the intrinsically disordered domain4 does not induce LLPS in the presence of the intrinsically disordered region. Together, these results suggest that the interaction between the intrinsically disordered region and the F-face of CBD is most important in the early stages of galectin-3-induced LLPS.
Note 4: One way to observe LLPS induction is to mix proteins and see if the mixture turns cloudy or if the clouding disappears reversibly with temperature changes or inhibitors. The glycoproteins used here were transmembrane glycoproteins. In this report, they used the soluble portions of these proteins, the ectodomain (the portion exposed outside the cell).
Then, does the interaction between the intrinsically disordered regions also contribute to LLPS/oligomerization of galectin-3? As described in Part 1, when galectin-3 binds to immobilized glycoproteins, it forms an oligomer and exhibits cooperative binding. Tai and Zhou’s group, therefore, used mutated galectin-3 (L203A) to examine cooperative binding on the T-cell surface. The intrinsically disordered regions of this mutant galectin-3 can bind to each other, but because of the mutation in the F-face of the CBD, the intrinsically disordered regions cannot bind to the CBD. Thus, by using this mutant, whether the interaction between the intrinsically disordered regions is important for LLPS/oligomerization can be studied. Cooperative binding was observed even with mutant galectin L203A at a concentration of 0.2 µM or higher, that is four times that of galectin-3. Therefore, whereas the interaction between the intrinsically disordered region and the CBD is necessary at the early stage of LLPS induction, the interaction between the intrinsically disordered regions is also involved once LLPS is induced or in the presence of high concentrations of galectin-3.
Tai et al.7 also produced 15 galectin-3 mutants in which specific prolines scattered in the first six repeats of the nine repeats of PGxY/W in the intrinsically disordered region were individually mutated to alanine and studied their activities and their potential for LLPS induction. Although there were differences in potency depending on the position of proline, many of the mutants showed decreased activity and decreased ability to induce LLPS. Thus, their scrutiny of the importance of prolines in the region suggests that the interaction between the intrinsically disordered region and the CBD is many to one, which is consistent with the inference by Huang et al. in 2017.
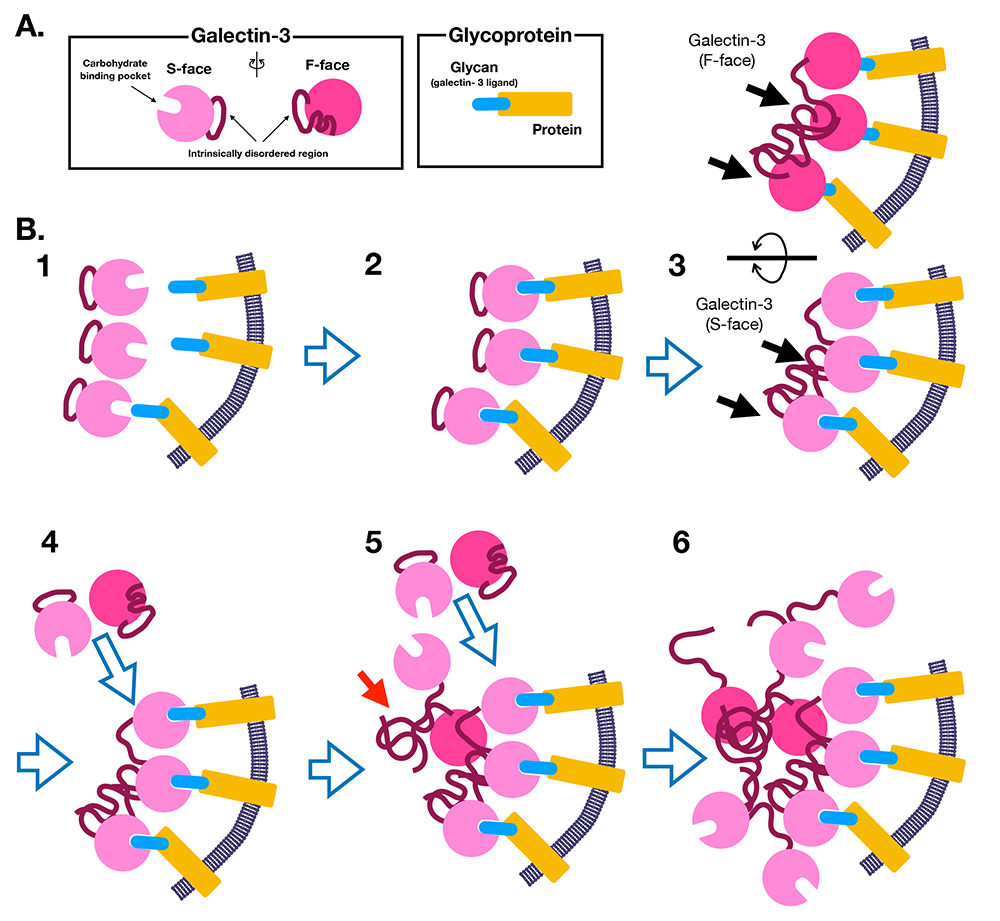
Based on three recent reports we discuss here on galectin-3 inducing LLPS, I would like to revise the molecular mechanism of galectin-3 oligomerization that I proposed in Part 1. LLPS-induced oligomerization of galectin-3 can be summarized as follows (Figure 5).
| 1) | The intrinsically disordered region of monomeric galectin-3 repeatedly interacts with and dissociates from the F-face of the CBD (intramolecular interaction) (Figure 5A and 5B-1). | 2) | Galectin-3 binds to specific ligands (e.g., glycoproteins) on the membrane (Figure 5B-2). | 3) | As the concentration and density of galectin-3 on the membrane increase and exceed the critical point for inducing LLPS, the intrinsically disordered regions begin to interact with the CBD of other galectin-3 molecules through intermolecular interactions (Figure 5B-3). | 4) | The concentration of galectin-3 on the membrane further increases, and the intrinsically disordered regions also oligomerize (Figure 5B-4, -5). | 5) | The local concentration of galectin-3 increases and oligomerization of galectin-3 is further promoted (Figure 5B-6). | 6) | The interactions involving the intrinsically disordered region are a dynamic and constantly repeating binding and dissociation. The molecules (galectin-3 and its binding partners) also reciprocally move and exchange with molecules outside the LLPS. |
Galectin-3 lattice formation is regulated and controlled at the biosynthetic level of its binding partners, glyans in various ways. For a detailed explanation, please refer to the Glycoforum by Dennis and Dimitriou11,12.
What about the control from the galectin-3 side? As previously mentioned, galectin-3-induced LLPS is a highly dynamic clustering maintained by interactions where the intrinsically disordered regions scan the F-face of the CBD that binds to glycans in addition to their oligomerization. It is worth noting that the intrinsically disordered region is highly sensitive to proteases. Some of the proteases reported to date include collagenase17, metalloproteases18, neutrophil elastase19, proteases from Staphylococcus aureus20 serine protease gp63 from the parasite, Leishmania21, and proteases from the Trypanosoma cruzi parasite that causes Chagas disease22. Since research on galectin-3 and LLPS has just begun, it has not yet been studied whether LLPS is affected in environments where these enzyms are present. However, considering that all the proteases mentioned here are capable of resolving the oligomerization through their high hydrolysis activity towards the intrinsically disordered region of galectin-3 and that the stability of LLPS depends on the length of the intrinsically disordered region, it can be speculated that the negative impact of proteases would be significant. For example, when neutrophils migrating to an inflammatory site become activated, degranulate, and secrete elastase, when necrotic neutrophils at an infected site release elastase, or when proteases of infecting microorganisms are present, it is likely that the nearby galectin-3-mediating LLPS would be rapidly dispersed.
In nature, cells have mechanisms to avoid the stochastic noise that occurs in biological reactions, but conversely, it is also thought that they have mechanisms that can utilize this type of noise to respond diversely. Because of various intrinsic and extrinsic factors and the inherent fluctuation unique to the biosynthesis of glycans, there is microheterogeneity in the glycan structures of glycoproteins. Sometimes, glycan microheterogeneity has biological significance, but other times it may be noise to be avoided or overcome23,24. To be bold, galectin-3-induced LLPS may cancel the noise level of glycan microheterogeneity. Because the focus of this forum is LLPS, I will not delve into discussions regarding galectin-3 glycan-binding specificity. Without fear of misunderstanding, it can be said that the galectin-3’s specificity for glycans is quite permissive compared with other galectins. First, in the case of the interaction of galectin-3 with purified glycans, for example, Galβ1-4GlcNAcβ1-3Galβ1-4GlcNA(Note 5), galectin-1 cannot bind to this structure once it is modified with α2-6 sialic acid (Siaα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc-). In contrast, galectin-3 can recognize this glycan since there is another lactosamine (Galβ1-4GlcNA) inside this glycan that is still intact (assuming it is not modified with α2-6). Moreover, the affinity of galectin-3 for glycans is proportional to the number of lactose residues on a single glycan. It is not particularly important whether the arrangement of these lactose residues on the glycan is parallel (e.g., three or four branches of complex N-glycans) or in tandem (e.g., polylactosamine)25,26.
Note 5: Gal: galactose, GlcNAc: N-acetylglucosamine, Sia: sialic acid
This ambiguous specificity of galectin-3 appears to be reflected in galectin-3-induced LLPS on cells. As Dennis’ group has already demonstrated in various systems, the regulation/control of the dynamic movement of membrane glycoproteins by the galectin-3 lattice is different depending on the number of glycans on the glycoproteins as well as the number of lactosamine residues11,12. Furthermore, Demetrious’ group has reported that immune regulation via the galectin-3 lattice was observed in the cells with impaired biosynthesis of highly branched complex N-glycan types. The compensation was achieved by increasing the biosynthesis of polylactosamines instead27.
Considering the high tolerance of binding specificity of galectin-3 for glycans, the microheterogeneity of glycans on membrane proteins may not have a significant impact on the early stage of galectin-3 lattice formation induced by galectin-3-initiating LLPS. The number and optimal density of lactosamines in the glycan cloud locally presented to galectin-3 may be more important, and slight structural differences in individual glycans may be canceled out by the LLPS process.
What is more critical could be the flexibility of the glycan structures invovled in LLPS, particularly how lactosamine residues are presented. For instance, bisecting GlcNAc restricts the conformation of the Manα1-3 and α1-6 arms of complex N-glycans, reducing the flexibility of glycans containing lactosamine. Although the presence or absence of bisecting GlcNAc does not seem to affect the affinity of galectin-3 toward glycans (personal communication with Dr. Jun Iwaki), it may have an impact during the induction of LLPS because it reduces the flexibility.
In addition, it may not always be optimal to have a densely packed glycan presentation. For example, there is a study by Percec’s group that examined how binding to galectins is altered by changing the density of lactose using self-aggregating glycondrimers as membrane mimics28. Unfortunately, only galectin-1 and -8 were used in the study, but the noteworthy finding is that galectin binding is higher with low density glycan dendrimers than with high density ones. In other words, it seems that lowering the glycan density suppresses the mutual interaction between glycans that occurs at high density, allowing more lactose to be efficiently recognized by galectin. We had reported that a similar occurrence may be occurring in galectin-329. gp120, a protein essential for HIV-1 infection of T-cells, is densely covered with glycans. Although it is known to be particularly rich in high mannose, several N-glycans are of the complex type, which are also clustered on the protein surface. Interestingly, galectin-3 readily binds to denatured gp120 but does not bind to intact (properly folded) gp120(Note 6). This also implies that, as described above, a moderate concentration of lactosamine is important for the induction of LLPS.
Note 6: Interestingly, galectin-1 binds to both intact nondenatured gp120 and HIV-1 viruses and increases the adhesion of HIV-1 to T-cells and the infection. In contrast, galectin-3 does not bind to the native HIV virus or gp120.
Given that galectin-3-induced LLPS may make the selection of galectin-3 binding partners ambiguous or fuzzy, other naive questions arise. How are membrane glycoproteins selected to enter the galectin-3 lattice?
There is an interesting report by Bertozzi and Weaver30. They demonstrated that synthetically produced bulky mucin-type glycolipid mimetics (80 µm height) could physically induce clustering (funneling) of integrins, membrane cell adhesion molecules (20 µm height). Furthermore, in cells that are forced to express bulky Muc-1 (200 µm height), the physical barrier of Muc-1 can guide (funnel) integrins to the matrix adhesion sites. The cytoplasmic domain of Muc-1 was not necessary for the clustering. Therefore, this report suggests the possibility that glycoproteins on the cell membrane are compartmentalized or partitioned according to their physical properties (such as bulkiness). The model system presented here is not a report of compartmentalization by lectins, whereas galectin-3 binds to both integrin and Muc-1.
It is thus possible that on the cell membrane, galectin-3-induced LLPS/lattice is not simply a single layer but can be physically partitioned into different layers according to the height (or bulkiness) of membrane glycoproteins, such as high layers (200 µm) like Muc-1 and low layers (20 µm) like integrins. The lattice formation and dissolution can be dynamically controlled by the local concentration of galectin-3 and interacting glycans and are involved in the function and endocytosis of glycoproteins present in the lattice at each layer level or partition. Indeed, Nabi’s group measured the behavior of N-cadherin and glycolipid GM1 regulated by galectin-3 lattice at the cell junction site by fluorescence recovery after photobleaching (FRAP)(Note 7) and reported that their behaviors were completely different. Based on this, they also speculated that the different behavior of the galectin-3 lattice containing N-cadherin compared with that containing GM1 may be due to the presence of different layers of lattice31.
Note 7: FRAP is a method for investigating the movement and dynamics of biological molecules. It involves focusing a beam of light on a specific region to cause fluorescence to disappear, followed by measuring the recovery of fluorescence as fluorescently labeled biomolecules move into the bleached area from the surrounding region. This allows for the measurement of the speed of movement and dynamics of the biological molecules.
Dennis’ group combined simulation and experimental results to show that the functional control of glycoproteins by the lattice as well as the control of the biosynthesis of highly branched glycans of these glycoproteins (via the availability of UDP-GlcNAc) is correlated with the number of N-glycans on each glycoprotein30. In other words, in the case of membrane proteins modified with a large number of N-glycans, the activity of the glycoproteins is sigmoidally controlled with respect to the availability of UDP-GlcNAc, which can increase the number of branchings of glycans. In the case of membrane proteins with a few N-glycans, the activity is controlled in an ON/OFF state. This report also suggests that the amount and density of lactosamine on the N-glycans of glycoproteins is the key to regulating the interaction between galectin-3 and membrane glycoproteins.
Most multitransmembrane proteins have only one or two N-glycans. In addition, many of these proteins are channels or transporters with little extracellular domain (ectodomain) outside the cell membrane. Therefore, it is anticipated that the galectin-3-induced LLPS lattice containing these glycoproteins resides in a low bulk layer near the cell membrane. From this perspective, although galectin-3-induced LLPS on the membrane occurs in 2-D, the lattices may have a 3-D spread with varying heights in different locations, considering the bulkiness and height of the glycoconjugates (glycoprotesins and glycolipids) that form the lattice.
As Dennis’s and Dimetriou’s groups explain in detail and discuss the roles of galectin-3 lattice in their Glycoforum11,12, the lattice formed by galectin-3-induced LLPS reduces the lateral (XY direction) diffusion velocity of membrane glycoproteins that are bound to galectin-3. This reduction in diffusion velocity can result in delayed clustering of receptor proteins necessary for signal transduction and slower endocytosis rates. Conversely, there are instances where galectin-3 oligomerization promotes the lateral movement and diffusion of glycolipids, such as GM1, and N-cadherin at cell–cell junctions but not elsewhere31. These findings suggest that regulation of endocytosis by ligand aggregation and clustering by galectin-3 work both in a suppressive and stimulatory manner.
A report by Johannes’ group on the role of galectin-3 in clathrin-independent carriers (CLICs) suggests that galectin-3-induced oligomerization regulates the behavior of membrane glycoproteins in a diverse manner depending on the concentration of galectin-333. Unlike clathrin-dependent endocytosis, CLIC endocytosis starts by forming tubular endosomal carriers with a unique morphology in which the membrane is invaginated and elongated. At the site of endocytosis, the formation of CLICs requires galectin-3 binding to glycoproteins (CD44, integrin α5 and β1), which are uptaken via CLIC. The process was shown to be dependent on the presence of glycolipids. However, it should be noted that not all proteins taken up via CLICs are galectin-3 dependent. Interestingly, when galectin-3 was artificially brought into proximity(Note 8) with glycolipids in model membranes containing lipids and glycolipids (but without any glycoproteins), the unique tubular endosomal carriers characteristic of CLICs were formed in a β-galactoside-dependent manner. Furthermore, it was revealed that the N-terminal intrinsically disordered region is necessary for this induction, indicating that galectin-3 oligomerization drives this endocytosis. This result suggests that binding of galectin-3 to glycoproteins is not essential for the formation of tubular endosomal carriers, but rather, it is imporatnt for galectin-3 to aggregate near glycolipids. Notably, this CLIC endocytosis can be induced at very low concentrations ranging from 0.3 nM to 33 nM, whereas at 330 nM, the CLIC endocytosis can be inhibited.
Note 8: By inserting nickel-containing lipids into the artificial membrane, His-tagged galectin-3 was forced to bind to the membrane.
Regarding this point, they further conducted research using a microcavity-suspended liquid bilayer model in which α5β1 integrin was inserted into another artificial membrane and reported it in 202234. In the classical biochemical experimental system described in my Glycoforum Part 1, cooperative binding of galectin-3 is observed at concentrations of 0.3 µM or higher, but in this highly sensitive system, the cooperative binding was observed even at 3.7 nM. At 3.7 nM, the aggregation of integrins with galectin-3 oligomer was small, and the diffusion rate increased. However, at concentrations of 18.5 nM or higher, lateral diffusion was divided into two types: a decrease and an increase in lateral diffusion, with an additional phenomenon of thickening of the membrane. The authors speculated that at low concentrations, galectin-3 oligomers bind to individual α5β1 integrins, whereas at high concentrations, this aggregation increases and forms a lattice.
From here, I can only describe it by imagination, but considering that galectin-3 and its ligand must reach a specific critical concentration for LLPS to occur, it is quite possible that the low-concentration phenomenon seen here is Step 3) of the formation mechanism of galectin-3 oligomer discussed in chapter 11), the state in which the intrinsically disordered regions interact with the CBDs of other galectin-3 (intermolecular interaction) (Figure 5B-3). At the early stage of the CLIC, it is difficult for galectin-3 to achieve stable interaction with glycolipids because of their low affinity for galectin-3. Thus, it may be the reason why galectin-3 on the membrane is first concentrated through binding to glycoproteins, and then galectin-3 may interact with glycolipids to induce CLIC. In Part 1, I discussed a study on the interaction between GM1 and galectin-335, and the size of the bound galectin-3 was not as large as when galectin-3 forms a lattice, and it was only less than 1.6 times the size of the CBD, which also supports this hypothesis. In contrast, at high concentrations, galectin-3 oligomerization may induce LLPS, leading to lattice formation. We very much look forward to future developments in the study of these highly interesting and seemingly opposing effects associated with galectin-3 oligomerization/lattice.
In the aforementioned 2022 report34, there are results suggesting that the intrinsically disordered region may actually interact with the lipid membrane. This may be due to the high hydrophobicity of amino acids, particularly tyrosine(Note 9), in the intrinsically disordered region. Ochieng’s group reported in 2005 that galectin-3 binds to phospholipids, galactosylceramide, and cholesterol immobilized on plates36. It is hoped that further research will shed light on how the hydrophobicity of this region affects the generation and dissolution of CLIC lattices.
Note 9: At 25°C, the amount of amino acids that can dissolve in 100 mL of water is 25 g for glycine, 0.51 g for asparagine, and only 0.054 g for tyrosine, which is lower than any other amino acid. In the intrinsically disordered region of galectin-3, eight tyrosine residues are relatively regularly scattered among the 106 residues.
As mentioned in Part 1, there are 15 galectins in humans, categorized into the prototype consisting of only a single CBD, the tandem repeat-type consisting of two CBDs connected through a relatively short peptide, and the chimeric galectin-3 with a unique intrinsically disordered region that is not found in any other animal lectins. So far, I have introduced and discussed papers on galectin-3 and LLPS. Now, I would like to discuss the possibility of the participation of other galectins in galectin-3-initiated LLPS.
In 2007, Hirabayashi’s group investigated the interactions of the tandem repeat-type galectin-9 with other galectins using surface plasmon resonance37. They reported that when galectin-9 was immobilized, it bound to galectin-3 or -8 but not galectin-1, and this binding was inhibited by lactose. Interestingly, when the N-terminal CRD of galectin-9 was immobilized, galectin-8 but not -3 binds, whereas immobilization of the C-terminal CRD resulted in the binding of galectin-3. These interactions were inhibited by lactose. If there is no steric hindrance, it is then possible that galectin-8 might bind to the N-terminal CRD of galectin-9 and galectin-3 binds to the C-terminal CRD.
The interaction sites between the CRDs of these different galectins remain unknown from this study. Because it is unknown, it is even more speculative, but if one of these sites is not AA200-220 on the F-face, which is the contact site with the intrinsically disordered region, would it not be possible that galectin-3 and -8 are recruited to galectin-9? Multiple galectins are produced and stored in the cytoplasm of many cells and tissues. Then, glycan-unbound galectin-3, -8, and -9 may form loose complexes in the cytosol. If such a formation happens, is there a possibility that this galectin complex would be first recruited as a complex to a site of action and then dissolved when one of them binds to the glycan, considering that these interactions are resolved by lactose? Galectins are also secreted via a leaderless secretory pathway or released from damaged or dead cells38. Because all galectins are likely secreted or released through the same pathway, it is also possible that they interact with each other in the extracellular environment. I look forward to the development of future research on this point as well.
So far, I have discussed and explored the LLPS induced by extracellular galectin-3. Galectin-3 is synthesized and stored in the cytosol, since it lacks a signal peptide sequence required for entry into the conventional secretory pathway38. Many reports have shown that intracellular galectin-3 is involved in various biological responses. Although not exhaustive here, I would like to discuss whether two biological responses inside cells, protection against lysosomal damage and RNA splicing, are related to LLPS.
Infection and other causes can damage the lysosomes inside the cell. For example, in the case of phagocytic cells of leukocytes, such damage occurs when microorganisms that have been phagocytosed and transported to lysosomes escape into the cytosol, rupturing the lysosome membrane. Various hydrolytic enzymes (e.g., proteases, glycosidases) leak out of damaged lysosomes into the cytosol and can cause further damage. Therefore, cells have a defense system called MERiT (membrane repair, removal, and replacement) against this type of damage39. When lysosomes are damaged, the glycoproteins on the lysosomal membrane are exposed to the cytosol. These exposed glycans are recognized by cytoplasmic galectins, and then membrane repair, removal of damaged lysosomes by autophagy, or generation of new lysosomes are induced depending on the degree of damage and the progress of the repair.
To elaborate in more detail39, first, galectin-3 is recruited to the exposed glycoproteins of the damaged lysosome, and MERiT is initiated. Galectin-3 bound to transferrin receptor forms a complex with proteins such as ALIX, a component of the endosomal sorting complexes required for transport (ESCRTs). This complex activates the ESCRTs mechanism, which is responsible for remodeling the membrane and facilitating membrane repair. In cases where repair by ESCRTs is difficult, the complex of lysosomal membrane protein, galectin-3, and E3 ubiquitin ligase, TRIM16 (tripartite motif-containing protein 16), modifies the damaged membrane with ubiquitin, inducing autophagy to remove the damaged lysosome. Furthermore, galectin-9 bound to lysosomal-associated membrane protein 2 (LAMP2) activates AMPK (AMP-activated protein kinase), which induces autophagy. In addition, this pathway inhibits the activation of mTOR, further promoting the removal of damaged lysosomes via autophagy. In the final stage of MERiT, galectin-8 binds to damage-exposed SLC38A9 and inhibits the activation of mTOR, activating the transcription necessary for lysosome replacement via TFEB. It is suggested that galectin-3 plays a more important regulatory role than other galectins in the early stages of the repair process of damaged lysosomes39.
Could galectin-3-mediated LLPS events occur on the damaged lysosome membrane? It has already been reported that LLPS is involved in autophagy40 Thus, although there is no paper yet, it can be inferred that galectin-3 induces LLPS in a similar way in which it occurs on the cell membrane. Another point to note is that the initiation of cellular defense mechanisms against damaged lysosomes is the recognition of lysosomal membrane proteins by galectin-3. As mentioned in the previous section, it has been reported that galectin-3 can form complexes with galectin-8 and/or -9, all of which are involved in this MERiT mechanism. Therefore, although it may be speculation, there is a possibility that the interaction between these galectins is involved in the cascade of defense mechanisms starting from the interaction between exposed glycans and galectin-3 within this galectin complex, followed by the release of galectin-8 and -9.
Lastly, I would like to touch upon the research on the interaction between galectin-3 and RNA, which started shortly after the gene sequence of galectin-3 was determined approximately 35 years ago41
In 1988, Wang’s group suggested that galectin-3 is one of the heterogeneous nuclear ribonucleoproteins (hnRNPs) involved in transcription and posttranscriptional modification (capping, splicing, polyadenylation) of pre-mRNA in the nucleus41. Subsequent studies using nuclear extracts showed that galectin-3 is involved in pre-mRNA splicing, and its activity can be inhibited by the N-terminal intrincisally disordered regions as well as by thiodigalactoside, an artificial sugar that blocks galectin-3’s glycan binding42,43. It is suggested that galectin-3 binds to U1snRNP (small nuclear ribonucleoprotein), which is involved in splicing, and is incorporated into the spliceosome, involved in pre-mRNA splicing44,45. However, this activity is not specific to galectin-3 since the prototype galectin-1 also exhibits this activity, whereas the deletion of galectin-3 (but in the presence of galectin-1) results in a different splicing pattern46.
In 2016, Jacob’s group confirmed that galectin-3 binds to 14 types of hnRNPs47. Furthermore, Pigny’s group suggested that galectin-3 is involved in the stability of MUC4 mRNA through hnRNP-L48. This effect is mediated by the galectin-3-dependent interaction of hnRNP-L with the CA repeat element of the 3'-UTR of MUC4, and thus has no effect on the stability of MUC1, which lacks this element. Pigny speculated that MUC4 may be stored in RNP granules along with galectin-3 because hnRNP-L is a constituent of RNP granules that store untranslated mRNA.
As described above, galectin-3 is involved in various aspects of RNA regulation, from posttranscriptional modification to stability. As mentioned briefly in my forum Part 1, membraneless organelles formed by LLPS inside cells often consist of RNA and proteins containing intrinsically disordered regions. Examples of such organelles include stress granules, P granules, and RNP granules. When galectin-3 is incorporated into hnRNPs, the intrinsically disordered region is not recognized by antibodies (such as Mac-2, which binds to AA48-100) against this region43. Considering that this region can inhibit splicing, it is possible that galectin-3’s involvement in RNA metabolism and incorporation into RNP granules also involves LLPS, which is dependent on the intrinsically disordered region in some way. Very recently, Wang’s group published a review on the relationship between galectin-3 and LLPS in the nucleus49.
Galectin-3 is a lectin with a wide range of biological activities, including nuclear, cytoplasmic, and extracellular functions. It is also the only mammalian lectin with a long intrinsically disordered peptide region. Research on galectin-3 and LLPS has only just begun. From an LLPS perspective, the interaction between galectin-3 and its binding partners is one-to-many and regulates the movement dynamics of its binding partners through the glycan-binding site of the CBD scanning the glycans with slightly ambiguous specificity and the intrinsically disordered region scanning the F-face and interacting with other galectin-3 intrinsically disordered regions. I hope that future studies on galectin-3, particularly those investigating its involvement in LLPS and its dynamic functions and regulations, will lead to further advances and contribute to a better understanding of the mechanisms behind its diverse functions.
I would like to express my deepest gratitude to Jim Dennis, who introduced me to essential papers (including the 2014 Rosen paper) and the concept of the Thompson problem and discussed with me on my previous forum. I also thank Dr. Yoshiki Yamaguchi for providing information on the flexibility of glycans, Dr. Jun Iwaki for sharing unpublished research on the binding of galectin-3 to bisecting glycans and Drs. John Wang and Yves St-Pierre for reading the manuscript and pointing out unclear points. Finally, I would like to thank Dr. Jun Hirabayashi for his thorough editing (especially for Japanese language editing).