
Toki Taira
Professor, Department of Bioscience and Biotechnology, Faculty of Agriculture, University of the Ryukyus, Japan
He obtained his M.D. and Ph.D. from Kyusyu University in 1998 and 2002, respectively. He was appointed Assistant Professor at the Department of Bioscience and Biotechnology, Faculty of Agriculture, the University of the Ryukyus in 1999 and was promoted to Associate Professor in 2006, and then Professor in 2015. He is presently engaged in the elucidation of the relationship between plant chitinases and their antifungal activities. Awards: Incentive Award of the Japanese Society for Chitin and Chitosan (2015), Okinawa Research Encouragement Award (2019), Technology Award of the Brewing Society of the Japan (2019), and Excellent Paper Award of the Society for Biotechnology, Japan (2021).

Tomoya Takashima
Assistant Professor, Department of Pharmacy, Faculty of Pharmacy, Osaka Ohtani University, Japan
B.S. (Agriculture), 2015, Kindai University, Nara, Japan
M.S. (Agriculture), 2017, Kindai University, Nara, Japan
Ph.D. (Agriculture), 2020, Kagoshima University, Kagoshima, Japan
Chitinases (EC 3.2.1.14) catalyze the hydrolysis of chitin, which is a β-1,4-linked homopolymer or oligomer of N-acetyl-D-glucosamine (GlcNAc). Many researchers accept that one of the physiological roles of these chitinases is to protect plants against fungal pathogens by degrading chitin, a major component of the cell wall of many fungi1,2. There is strong correlative evidence that low constitutive activity of chitinase found in many plants can be dramatically induced by infection with fungal pathogens. And chitinases with antifungal activity have been reported in bacteria3,4. Not all chitinases exhibit antifungal activity, and rather few chitinases with strong antifungal activity have been reported. In this review, we will summarize what is known so far about the relationship between the structure and antifungal activity of chitinases, focusing on the results of our research. Elucidation of the correlation between chitinase structure and antifungal activity is expected to contribute to the understanding of plant defense systems, breeding of pathogen-resistant crops, and development of antifungal agents.
According to the CAZy database (http://afmb.cnrs-mrs.fr/CAZY/), there are two families of chitinases: glycoside hydrolase (GH) family 18 (GH18) and GH family 19 (GH19). GH18 and GH19 chitinases differ not only in sequence but also in hydrolytic mechanisms because they operate by retention and inversion, respectively, of the anomeric configuration5.
The first three-dimensional structure of a GH19 catalytic domain elucidated, the GH19 catalytic domain of barley GH19 chitinase has five internal loop regions and a C-terminal loop region (Fig. 1)6. Some GH19 chitinases have one or more deletions. These deletions are restricted to loop regions and the C-terminal loop region. There are variations in the number of deletions and their regions. PLC-A7, pokeweed leaf chitinase-A (Q7M1Q9), lacks loops II, IV, V, and the C-terminal loops; BcChi-A8,9, Bryum coronatum chitinase-A (BAF99002), lacks all but loop III, and pathogenesis-related proteins (PR-proteins) from tobacco, PR-P and PR-Q10, lack only loop III (Fig. 1). The bacterial chitinases Streptomyces griseus chitinase-C11 (Sg-ChiC) and Streptomyces coelicolor chitinase-G12 (Sc-ChiG) lack loops I, II, and V as well as C-terminal loops (Fig. 2).
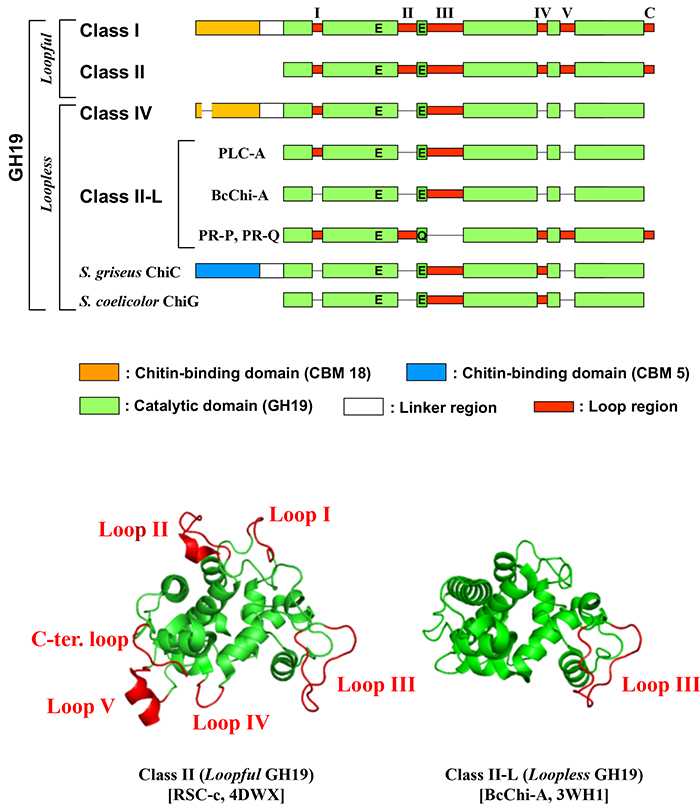
Ohnuma et al. defined "loopful" GH19 chitinases as those without loop defects and "loopless" GH19 chitinases as those with defects in the loop region (Fig. 1)13. Ohnuma et al. have determined the complex structure of rye seed chitinase RSC-c, a "loopful" GH19 chitinase, and BcChi-A, a "loopless" GH19 chitinase, with chitin oligosaccharides9,13. The "loopful" GH19 chitinase has a subsite structure with -4 to +4 and the "loopless" GH19 chitinase has a subsite structure with -2 to +2. Loops of the "loopful" GH19 chitinase are involved in substrate binding, doubling the length of the substrate recognition cleft of the "loopless" GH19 chitinase. Due to the extended substrate binding cleft, the tetrasaccharide chitin oligosaccharide is less likely to bind to the "loopful" GH19 chitinase across the active center, and the rate of tetrasaccharide degradation is about 1/1000 of that of the "loopless" GH19 enzyme8.
Some GH19 chitinases have a carbohydrate-binding module (CBM)5. In plants, GH19 chitinases have a chitin-binding domain attached to their N-terminus; the chitin-binding domain belonging to CBM18 is homologous to hevein, an antifungal peptide from rubber latex14,15. Bacterial GH19 chitinases have mainly CBM5/1211 or fibronectin type III domains16.
Plant GH19 chitinases are classified by their domain combination and presence or absence of loop(s) in the catalytic domain. Class I chitinase consists of a CBM18 and "loopful" GH19 catalytic domain; class II chitinase consists of a loopful GH19 catalytic domain only15; class IV chitinase consists of a CBM18 with a deletion and "loopless" GH19 catalytic domain17; class II-L chitinase consists of "loopless" GH19 catalytic domain only8.
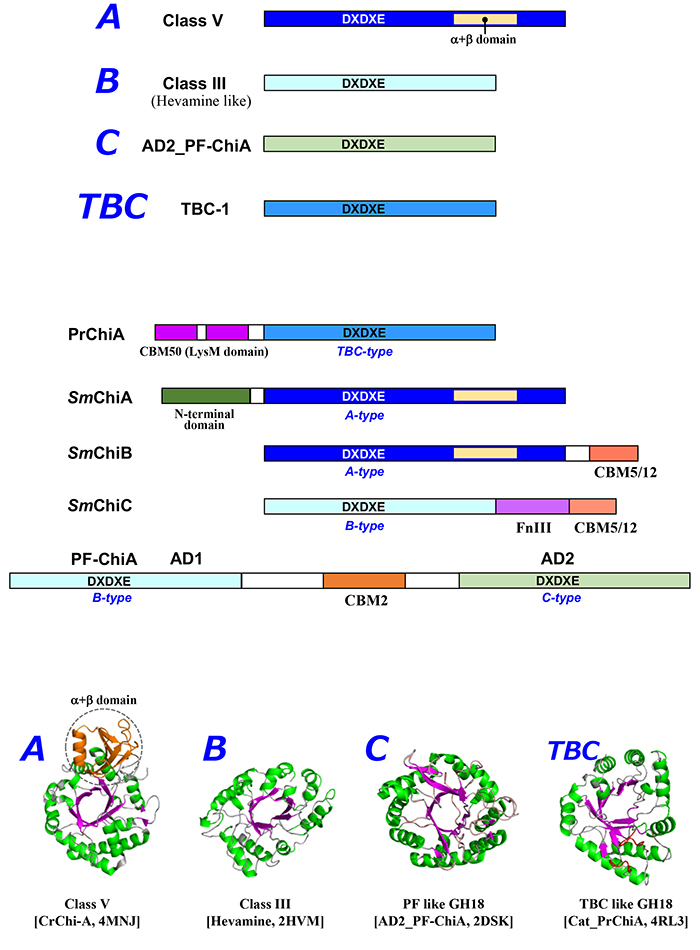
The 30-kDa chitinase hevamine18 from the Para rubber tree, the first GH18 chitinase to be structurally characterized, has the simplest (β/α)8-barrel structure and is in class III of the plant class classification15. GH18 chitinase shares the (β/α)8-barrel structure with the consensus motif DxDxE around the catalytic cleft, but the molecular structure of chitinase varies widely. Yamagami et al19. reported that tulip bulb chitinase (TBC) has low sequence homology (<15%) to Class III chitinases such as hevamine. TBC has no disulfide bonds, while class III chitinases have three conserved disulfide bonds; TBC has homology to narbonin (plant 2S protein from the globulin fraction of narbon bean (Vicia narbonensis), and the chitin-oligosaccharide degradation pattern of TBC is different from class III chitinases. The GH18 catalytic domain of Pteris ryukyuensis chitinase-A (PrChiA)20, which shows high homology to TBC, contains two large loop regions not found in hevamine, suggesting a different mode of action on the substrate21. Because of this significant difference in structure and properties, the TBC-like GH18 chitinase is considered to be in a separate class from class III. The 40-kDa GH18 chitinase from tobacco has an additional (α+β) domain consisting of about 70 amino acid residues in the middle of the (β/α)8-barrel structure (Fig. 1)22. The (α+β) domain serves as a "wall" on one side of the substrate binding cleft, making it the substrate binding site of the enzyme deep cleft. Therefore, the structure of the substrate-binding subsite and its sensitivity to inhibitors is very different from that of GH18 class III chitinase. This type is class V in the classification of plant chitinase23.
Bacterial GH18 chitinases have been classified into subfamilies A, B, and C by Watanabe et al24. Fungal GH18 chitinases have been classified into subgroups A, B, and C by Seidl et al25. Subfamilies A and B of bacterial GH18 chitinases correspond to fungal subgroups A and B. However, subfamily C of bacterial GH18 chitinases is quite different from fungal subgroup C. Fungal subgroup C consists of CBM18 and CBM50 linked to the GH18 catalytic domain, which is relatively homologous to bacterial subfamily A25. To avoid confusion, this review rests on a classification of bacteria that is based on homology of the catalytic domain only: A, B, C, and other "types" (Fig. 2). Type A is considered the bacterial/fungal type because it is common in bacteria and fungi. Microbial A-type GH18 has a tobacco GH18 chitinase-like structure with an additional (α+β) domain between the seventh and eighth β-strands of the (β/α)8-barrel structure and is characterized by a deep substrate binding cleft. Therefore, this type is homologous to plant class V and is also considered class V type. The catalytic domain of this type is also homologous to human chitotriosidase. Type B is considered the fungal/plant type because it is more common in fungi and plants. It has a simple (β/α)8-barrel structure similar to hevamine, and is therefore also considered a class III type. Type C was first structurally identified in the archaeon Pyrococcus furiosus (PF-ChiA)26, and homologous GH18 chitinases have been obtained from bacteria and eukaryotes such as Euglena27 TBC-like GH18 chitinases are in a distinctly different group from types A, B, and C. According to classification by the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/)28, the GH18_chitinase-like superfamily (cl10447) is divided into 17 families. The phylogenetic tree based on these data supports the above classification (Fig. 3).
In summary, it is suggested that the GH18 chitinase catalytic domain is divided into type A (Fungi/bacteria, class V-like), type B (Fungi/plants, class III-like), type C (PF-ChiA-like), type TBC (narbonin-like), and other chitinases throughout the biological world.
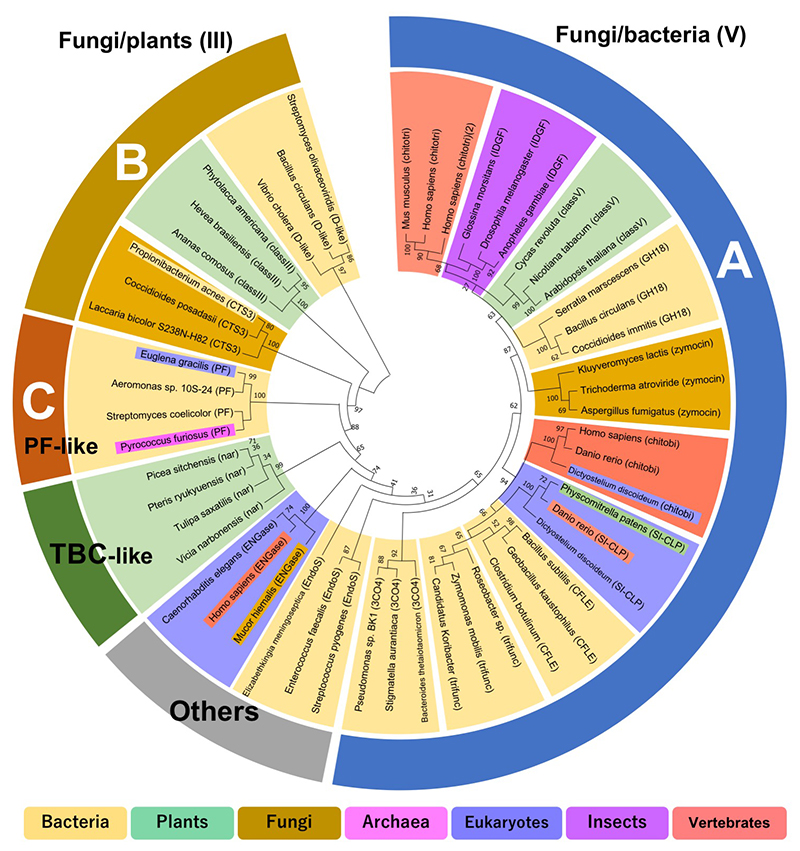
Comparing papers on the antifungal activity of GH18 and GH19 chitinases, there are clearly more reports of the antifungal activity of GH19 chitinase. Beginning with GH19 chitinase from bean29, which was the first chitinase to show antifungal activity, most of the reports of antifungal activity of chitinases have been about GH19 chitinases from legumes, wheat, tobacco, and other sources30. Watanabe et al. reported that GH19 chitinase ChiC from Streptomyces griseus showed antifungal activity against Trichoderma reesei, but the GH18 chitinase ChiA1 from Bacillus circulans WL-12 and Serratia marcescens ChiA, which has high chitinolytic activity, did not show antifungal activity3. Kawase et al. reported that Chi19F, a GH19 chitinase from Streptomyces coelicolor A3(2), showed antifungal activity against Trichoderma reesei, while GH18 chitinase from the same strain showed no activity4.
In regard to plant-derived GH18, it was reported that class V chitinase from tobacco exhibits antifungal activity31. Onaga and Taira showed that PrChiA, consisting of a TBC-like GH18 catalytic domain and two LysM domains (CBM50), inhibits the growth of Trichoderma viride20. Deletion of the two LysM domains of PrChiA abolished its antifungal activity.
There have been several reports on the contribution of chitin hydrolysis activity in the exertion of antifungal activity of chitinase. The mutant protein of class I chitinase from chestnut seeds that had no chitinolytic activity caused as much morphological change of T. viride hyphae as the wild-type chitinase32. The mutant protein of the 26-kDa chitinase (class II) of barley that does not possess chitinolytic activity had 15% of the antifungal activity of the wild-type chitinase33. Ohnuma et al. found that a chitinolytically inactive mutant of class I chitinase from rye seed maintained chitin-binding activity but exhibited no antifungal activity at all34. Gazyumaru latex chitinase-B (GlxChiB, GH19 class I chitinase), which has strong antifungal activity, lost its antifungal activity in the chitinolytically inactive mutant35. The chitin-degrading inactive mutant of PrChiA showed no antifungal activity20. It is true that some chitinase-mutants having no chitin-hydrolytic activity exhibit slight antifungal activity, however, there is no doubt that the antifungal activity of most chitinases stems from their chitinolytic activity.
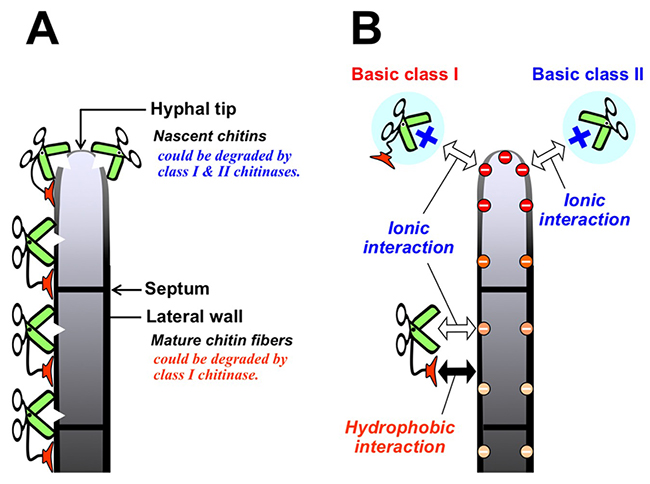
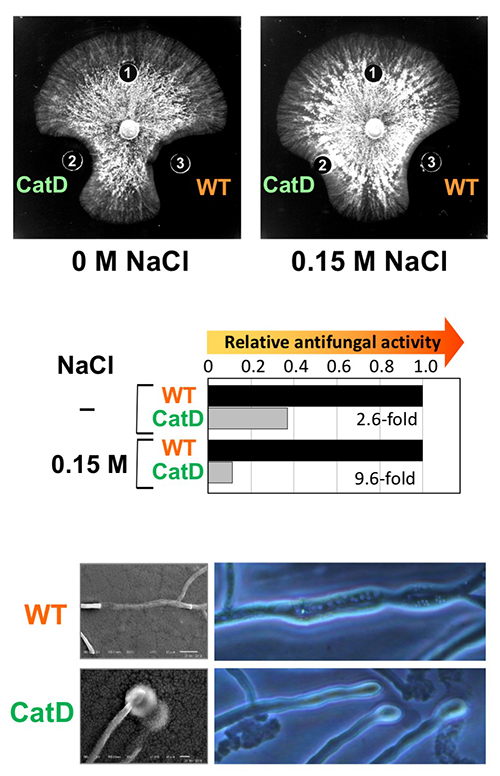
In several types of plant chitinases, class I chitinase is of interest due to the role of the chitin-binding domain in antifungal activity. Hevein14 and Ac-AMP36 (antimicrobial peptide from Amaranthus caudatus seeds), which have homology to the CBM18 chitin-binding domain of class I chitinase, exhibit antifungal activity. The chitin-binding domain of class I chitinase from rye seeds had no or very low level of antifungal activity, whereas the catalytic domain of this chitinase exhibited the same level of antifungal activity as that of class II chitinase37. Class II chitinase derived from barley seed inhibited the growth of the fungus T. viride to the same extent as class I chitinase did38. Wild-type class I chitinase and chitin-binding domain-deletion mutants of class I chitinase from tobacco inhibited T. viride growth, but the wild-type chitinase was more potent than the mutants39,40. Using a unique bioassay, Taira et al. showed that class I chitinases clearly inhibit mycelial elongation better than class II chitinases41. These reports suggest that the chitin-binding domain of class I chitinase strongly contributes to the antifungal activity of this enzyme. During apical growth in filamentous fungi, chitin and β-glucan fibers are synthesized simultaneously in the tip of the growing hypha. In the fungal mature cell walls, at a distance from the hyphal tip, the polysaccharides are cross-linked to form mixed chitin-glucan fibers and may be overlaid by other polysaccharides and protein layers (Fig. 4A). Taira et al. showed that FITC-labeled class I chitinase bound to the hyphal tips, lateral walls, and septa but FITC-labeled class II bound only to the hyphal tip. Furthermore, class I had a greater affinity for the cell walls than class II. Class I chitinase liberated a larger amount of reducing sugar from the cell walls than class II did. These results inferred that class I chitinase binds to the lateral walls and septa of mature cells and degrades mature chitin fiber, while class II binds only to the hyphal tip followed by degradation of only nascent chitin (Fig. 4A). By real-time observation using optical microscope and SEM images, Takashima et al. showed that class I chitinase (GlxChiB) acts not only on the hyphal tips of T. viride but also on the lateral walls of the hyphae, whereas its CBM18 deletion mutant acts only on the hyphal tips (Fig. 5)35.
In bacteria, many GH19 chitinases are CBM5 (ChtBD3 superfamily) linked. Wild-type ChiC from S. griseus, consisting of CBM5 and GH19, exhibited antifungal activity against T. reesei, while CBM5 deletion mutant showed almost no antifungal activity42. ChiB from Nocardiopsis prasine, which has a domain combination similar to that of ChiC, exhibited antifungal activity against T. reesei in the wild type, but greatly reduced antifungal activity in the CBM5 deletion mutant43. GH19 chitinase (CHI1) from Bacillus circulans KA-304 has an FnIII-like domain with chitin-binding ability at its N-terminus, and its deletion caused a marked decrease in protoplast formation16. The contribution of CBM is significant in the antifungal activity of GH19 chitinase in bacteria.
There are many reports about the antifungal activity of plant GH19 chitinases, however, more of them are about the antifungal activity of basic isozymes than of acidic isozymes30. Basic class I chitinase from pineapple leaves showed strong antifungal activity, while acidic class I chitinase had very weak antifungal activity44. Ohnuma et al. showed that basic class I chitinase from rye seed can maintain high antifungal activity under high-salt conditions (on medium supplemented with 0.15 M NaCl), whereas the catalytic domain alone exhibits significantly reduced antifungal activity under high-salt conditions34. Chitin-binding activity of GlxChiB (basic class I chitinase)was enhanced under high-salt conditions45. The chitinase keep high antifungal activity under high-salt conditions, whereas the antifungal activity of the chitin-binding domain deletion mutant was significantly reduced under high-salt conditions (Fig. 5)35. Acidic class I chitinase from pineapple leaves showed antifungal activity under high-salt conditions, but not under low-salt conditions44. These findings suggest that basic class I chitinase binds to the fungal cell wall by both hydrophobic interaction of the chitin-binding domain and ionic interaction of the catalytic domain (Fig. 4B). Taira et al. showed that basic class II chitinase bound to a column packed with fungal cell wall fraction prepared from the mycelia of Trichoderma sp., under pH 6.0 and low ionic strength conditions34. The binding ability of basic class II chitinase was reduced by elevating pH or ionic strength. Many antimicrobial peptides have high basicity, which would be sufficient for antifungal activity36. The surface of microbial cells is negatively charged due to negatively charged phospholipid headgroups. Basic chitinases might bind to the fungal cell surface by ionic interaction2.
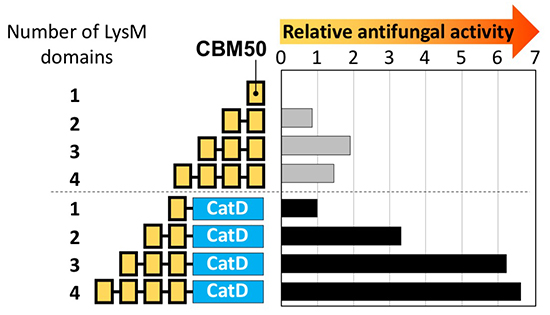
Takashima et al. showed that multimers of the second LysM domain from the N-terminus of PrChiA exhibit antifungal activity (Fig. 6)46. Not LysM monomer but LysM multimers (LysMn, n=2–5) showed antifungal activity, with LysM3 being the most active. Not the catalytic domain alone, but multimer fusion chitinases (LysMn-Cat, n=1–4) exhibited antifungal activity, and LysM3-4 fused chitinases showed the highest activity. Microscopic observation revealed that LysM multimers attacked only the tips of the fungal hyphae, while LysM fused chitinases attacked the tips and the lateral walls around the septa of the fungal hyphae.
In this review, I presented the structure of chitinase and the role of each domain in its antifungal activity. While some fungi are highly sensitive to chitinase (e.g., Trichoderma spp.), many strains are not susceptible or not inhibited by chitinase alone. This may be due to the fact that the fungal cell wall is composed not only of chitin but also other polysaccharides and glycoproteins.
Plants have developed defense mechanisms to prevent fungal infection, and fungi have developed mechanisms to infect plants. The cell wall is the front line of contact between fungi and plants, and both have evolved enzymes to degrade the others’ cell walls, while at the same time evolving their own cell wall structures (Fig. 7). The co-evolution continues, with some fungi having α-1,3-glucan as a cell wall component commonly accompanied by chitin and β-1,3-glucan47. Plants have enzymes for chitin and β-1,3-glucan degradation and receptors for detection of them as defense mechanisms, but lack enzymes and receptors for α-1,3-glucan. It has been reported that α-1,3-glucan on the fungal cell wall allows some fungi to bypass the plant's biological defense against infection48. Bacteria also have fungal cell wall degrading enzymes such as chitinases and β-1,3-glucanases to compete for supremacy with fungi and/or to intake fungal cell walls. Some bacterial species have α-1,3-glucanases49 and β-1,6-glucanases50, but these have not been found in plants.
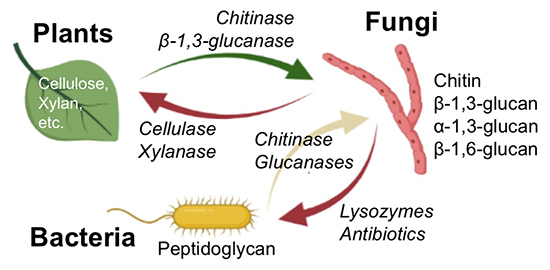
Currently, there is a need for safe and effective antifungal agents in various fields, including medicine. As with antibiotics (anti-bacterial agents), the use of small-molecule antifungal drugs has led to the emergence of drug-resistant strains. In principle, resistance to fungal cell wall degrading enzymes is expected to be very limited. Based on the above, we believe that the development of cell wall polysaccharide-degrading enzymes as antifungal agents is valid. To construct such a novel antifungal system, it is necessary to study not only plant-derived chitinases and β-1,3-glucanases, but also various fungal cell wall polysaccharide-degrading enzymes of bacterial origin for their combined antifungal activity. Chitinase should play a central role in the antifungal system since chitin is the predominant fungal cell wall component.