
John L. Wang
John L. Wang received his Ph.D. degree from The Rockefeller University in 1973. In 1977, he joined the faculty in the Department of Biochemistry and Molecular Biology, Michigan State University, where he is now Professor emeritus. He has had a long-standing interest in lectins: (a) He worked on determining the amino acid sequence and X-ray structure of the plant lectin, concanavalin A (Con A). An analysis of the inhibition by Con A of the mobility of cell surface receptors on lymphocytes provided one of the early lines of evidence for receptor-cytoplasmic/cytoskeletal interactions. (b) His laboratory also investigated the binding and adhesion of the bacterium Bradyrhizobium japonicum to roots of soybean plants leading to a nitrogen-fixing symbiosis. This analysis led to the discovery that the galactose-specific binding of rhizobium to soybean cells is mediated by a lectin present at one pole of the Bradyrhizobium japonicum cell. (c) Finally, he and colleagues have reported on the phenomenon of dual localization of galectin-3, in the extracellular compartment as well as in the intracellular compartment. In the cell nucleus, galectin-3 is co-localized with other pre-mRNA splicing factors in nuclear speckles, which are membrane-less organelles formed by the phenomenon of liquid-liquid phase separation.
Galectins are saccharide-binding proteins with characteristic amino acid sequences in the carbohydrate recognition domain. Many members of the galectin family also show two other features: they reside both inside as well as outside of cells and they bind multiple partners through protein-protein interactions. Inside cells, some examples of non-carbohydrate ligands of various galectins include: (a) prototype galectin-1 binds to oncogene H-Ras and transcription factors OCA-B and TFII-I; (b) tandem-repeat type galectin-8 binds to other galectins such as galectin-9, the autophagy receptor NDP52, and tripartite motif protein TRIM5α; and (c) the NH2-terminal domain of chimera type galectin-3 binds to components of the endosomal sorting complex required for transport such as Tsg101 and to ribonucleoprotein complexes such as hnRNP A2B1 while the COOH-terminal domain binds to the apoptosis repressor Bcl-2, tripartite motif protein TRIM16, and transcription factors OCA-B, TFII-I, and β-catenin. The present article aims to summarize selected studies that highlight the promiscuity of intracellular galectins and several intriguing questions raised by these findings.
Galectins are a family of animal lectins defined on the basis of binding affinity for β-galactosides and conserved sequence elements in the saccharide-binding site1. As of this writing, a PubMed search using the keyword “galectin” would yield nearly 11,000 articles on some 16 members of the family. These studies have shown that galectins are involved in many physiological and pathological processes. At least part of the reason for the large number of studies has to do with the fact that galectins exhibit a diverse range of subcellular localizations and molecular interactions. Many members of the galectin family have been found both in the extracellular compartment (cell surface and medium) as well as in the intracellular compartment (cytoplasm and nucleus)2. In the intracellular compartment, the activities of various galectins are often classified as either glycan-dependent (based on inhibition by saccharide ligands) or glycan independent. However, even those activities that are inhibited by carbohydrate ligands could involve binding interactions distinct from the saccharide-binding site.
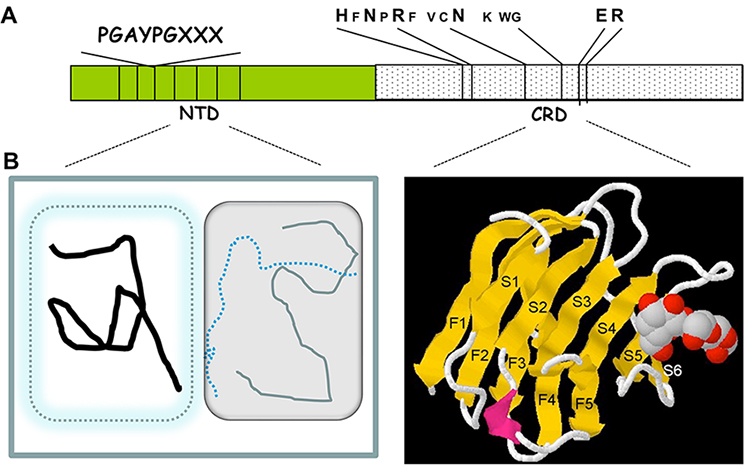
The present discussion in the Glycoforum on galectins aims to highlight that intracellular galectins are highly promiscuous, binding to multiple partners. I will use galectin-3 (Gal3) as the prime example but I hope to interject comparisons with other galectins to which many investigators have contributed interesting findings. In terms of content and organization of domains, Gal3 is the sole representative of the chimera subgroup of the galectin family3. The Gal3 NH2-terminal domain (NTD) is characterized by internal sequence homologies containing a repeating motif rich in proline and glycine residues4; the carboxyl-terminal half shows sequence similarity with the corresponding carbohydrate recognition domains (CRD) of other members of the galectin family, with highly conserved amino acid residues that interact with saccharide ligands as revealed by X-ray crystallography5 (Fig. 1A and 1B). Because of the promiscuity, both by the homologous CRDs in different galectins and by the intrinsically disordered NTD of Gal3, galectins can serve as platforms for assembly of a multitude of macromolecular complexes (Tables 1 and 2).
The NTD of Gal3 (residues 1 to about residue 137) exhibits low amino acid sequence complexity, with multiple repeating motifs rich in proline and glycine; one example of alignment displays a 9-residue repeating motif Pro-Gly-Ala-Tyr-Pro-Gly-Gln-Ala-Pro, with some substitutions (hereafter designated as the PGAYPG motif) (Fig. 1A). Computational algorithms for prediction of disordered regions such as IUPred8 or PONDR9,10 suggest that the NTD is an intrinsically disordered region (IDR) while the CRD folds like a classical globular protein (Fig. 1B). Indeed, UV-CD spectroscopy, NMR and nuclear Overhauser effect spectroscopy (NOESY), and differential scanning calorimetry all indicate that the Gal3 NTD is largely unstructured11-13. On the other hand, these naturally unstructured regions of polypeptides are highly flexible, are notoriously promiscuous in terms of binding to multiple partners, and can assume a “more ordered” conformation upon interaction14. In accord with the suggestion6 that new visual representations be developed to accommodate the special features of IDR in polypeptides, especially in terms of conformational heterogeneity, the NTD of Gal3 is displayed in Fig. 1B as a composite of different depictions.
Barboni and co-workers15 as well as Berbís et al.16 used molecular modeling, mutagenesis, and synthetic peptide titration studies to document evidence that the NTD of Gal3 interacts with sequences in its own CRD. The latter group also showed transient secondary helical conformation and proline-mediated multi-turn structures around the PGAYPG repeats17. Finally, Lin et al.8 showed that the region of the Gal3 NTD containing the PGAYPG motifs are involved in interactions, both with another NTD or with the CRD (Tables 1 and 2). Intermolecular NTD—NTD and NTD—CRD interactions provide one mechanism of Gal3 self-association to form oligomers18-21, as suggested by observations such as agglutination of erythrocytes22,23 and positive cooperativity in the binding to multivalent glycoconjugates23,24. The ability to oligomerize for multivalent binding, along with the IDR in its NTD, allows the Gal3 polypeptide to undergo the physico-chemical process of liquid-liquid phase separation (LLPS). Huang and co-workers first reported LLPS of purified preparations of full-length Gal3 and its NTD and carefully analyzed its dependence on protein concentration, ionic strength, temperature as well as its reversiblity8,9.
In addition to interacting with its own NTD and CRD, the IDR of Gal3 can also bind to bacterial lipopolysaccharide (LPS)25 (Table 1). Fermino et al. documented that LPS-binding induces Gal3 oligomerization and enhancement of neutrophil activation20. This is but one example of NTD-mediated aggregation, resulting from multiple weak interactions amongst Gal3 molecules that “agglutinate” LPS micelles9. More generally, Zhao et al. showed that Gal3 binding to cell surface glycans resulted in phase separation, which, in turn, affected outcomes such as T-cell activation, etc.26. The role of LLPS in Gal3 binding to cell surface glycoconjugates has been the subject of two prior Glycoforum discussions27,28; the present focus on intracellular galectins will discuss LLPS of Gal3 in intracellular membraneless organelles (see Section 3 below).
Tsg101 is one component of ESCRT (endosomal sorting complex required for transport), involved in a number of processes including the formation of multivesicular bodies, retrovirus budding, and the repair, removal and replacement of damaged endolysosomal membranes. Tsg101 interacts directly with Gal3 (Table 1) through recognition of a tetrapeptide motif P(S/T)AP (found, for example, in residues 74—77 and residues 83—86 amongst the PGAYPG repeats of murine NTD)29. This Tsg101—Gal3 interaction is responsible for recruitment of Gal3 into the lumen of exosomes which, in turn, accounts for the release of Gal3 into the extracellular space. Indeed, knockdown of Tsg101 or mutation of the P(S/T)AP sequence decreased exosomal secretion of Gal329. When the NTD of hamster Gal3 was fused onto the normally cytosolic protein chloramphenicol acetyltransferase (CAT), the fusion protein was secreted from transiently transfected Cos cells30. Systematic truncation of the hamster NTD sequence identified 89YPSAPGAY96 as critical for secretion. Significantly, it was pointed out that the short sequence of residues 89—96 in hamster Gal3, by itself, was not sufficient to direct the secretion; that sequence needed to be recognized within the context of the PGAYPG repeats in the NTD30.
A yeast two-hybrid screen of a Jurkat T lymphoma cDNA library using Gal3 as a bait yielded another ESCRT component Alix (ALG-2 interacting protein) as a binding partner31 (Table 1). The interaction of Gal3 with ESCRT components Alix and Tsg101 is also important in the repair of small disruptions in endolysosome membranes. Gal3 recognizes glycans exposed, including the glycosylated protein TFRC, when there is endolysosomal damage such as seen during infection by Mycobacterium tuberculosis. Gal3 then recruits Alix and facilitates the interaction of the latter with downstream ESCRT-III effector CHMP4 in the process of restoring lysosome function32. Radulovic et al.33 reported that CHMP4B accumulated at damaged lysosomes much earlier than Gal3, suggesting that ESCRT-III components detected a subtler membrane damage than Gal3 recognition of exposed intraluminal glycans. Nevertheless, it is important to note that the CRD of Gal3 binds to TRIM proteins (see Section 5 below on the non-carbohydrate ligands of the CRD). Therefore, Gal3 can switch from NTD-mediated ESCRT interactions (e.g. Alix and Tsg1010), involved in membrane repair33, to CRD-mediated TRIM interactions (e.g. TRIM16), involved in autophagic removal of damaged lysosomes32.
| Liganda | Inhibition by saccharidesb | Gal3 NTD | Noc | Gal3 CRD | No | Lipopolysaccharide | No | ESCRT component ALIX | No | ESCRT component Tsg101 | Not directly tested | hnRNP A2B1 | Yes | hnRNP-L | Not directly tested | PSF RNP complex | Not directly tested | U1 snRNP | Yes |
|---|
a NTD, NH2-terminal domain of galectin-3 (Gal3); CRD, Carbohydrate recognition domain.
b Experiments to determine sensitivity to inhibition by saccharide ligands of the carbohydrate recognition domain were carried out with full-length Gal3.
c Intermolecular NTD—NTD interactions are facilitated when the CRDs of some Gal3 molecules are bound to saccharide moieties of multivalent glycoconjugates (e.g. laminin; lipopolysaccharide), increasing the local Gal3 concentration. Lactose dissociates the CRD binding to multivalent glycoconjugates, thereby disrupting the aggregates.
Fritsch et al.34 documented that Gal3 binds to hnRNP (heterogeneous nuclear ribonucleoprotein) A2B1 (Table 1) and co-localizes with the latter in punctate structures in the nucleus. This was carefully documented by in situ proximity ligation assay between the two proteins and between hnRNP A2B1 and SC35. Consistent with these results, the colocalization of Gal3 and SC35 has been reported35. The SC35 monoclonal antibody, which recognizes an epitope on Serine/Arginine Repetitive Matrix Protein 2 and members of the SR family of splicing factors, is a signature of nuclear speckles/interchromatin granule clusters36. Nuclear speckles are one of a number of morphologically distinct substructures, collectively known as nuclear bodies, including the nucleolus, coiled bodies and gems, and promyelocytic leukemia (PML) bodies. These are not membrane-bound structures but arise from LLPS37. The association of Gal3 with SC35 in nuclear speckles indicates that Gal3 in the nucleus participates in LLPS under conditions endogenous to the cell. Gal3 possesses three key features for its localization in liquid-phase condensates: (a) an intrinsically disordered domain; (b) oligomer formation for multivalent binding; and (c) association with RNA and ribonucleoprotein complexes10.
Nuclear speckles contain various hnRNPs and small nuclear ribonucleoprotein particles (snRNPs) as well as a host of other splicing factors36. Indeed, depletion and reconstitution experiments have documented that Gal3, as well as galectin-1 (Gal1), are functionally redundant components of the splicing pathway35,38-40. Moreover, HeLa cells transfected with Gal1- and Gal3-specific siRNAs to knockdown the expression of the lectins yielded changes in patterns of mRNA splicing and transport, as revealed by RNA-seq analysis34. In the absence of a pre-mRNA substrate, the polypyrimidine tract binding protein-associated splicing factor (PSF) forms a large complex containing five snRNPs and many splicing factors, including Gal341. In the canonical model of step-wise assembly of the spliceosome, the pre-mRNA is initially complexed with hnRNPs to form the so-called H-complex; addition of U1 snRNP in the absence of ATP results in an E-complex committed to the splicing pathway42. In the presence of ATP, various other uracil-rich snRNPs are added to form the active spliceosome. It appears that Gal3 actually gains entry to pre-mRNA splicing through association with U1 snRNP (Table 1), in the first step formation of E-complex39,40.
A synthetic peptide bearing three repeats of the PGAYPGQAP sequence (27-mer corresponding to residues 41—67 of the murine sequence) inhibited the splicing reaction43. In contrast, a mutant 27-mer synthetic peptide with the same amino acid composition but with a scrambled sequence showed no inhibitory effect. Finally, purified preparations of recombinant murine NTD also inhibited H-/E-complex formation and the splicing reaction whereas purified preparations of the CRD did not44. These results document the dominant negative effect of the PGAYPG motif on splicing and suggest that the intrinsically disordered NTD contributes to the interaction of the Gal3 polypeptide with the splicing machinery.
Finally, Gal3 binds to hnRNP-L (Table 1) and, through the latter’s interaction with the CA-repeat elements in the 3’-untranslated region of the mature MUC4 mRNA, stabilizes that mRNA45. In contrast to the nuclear bodies discussed above, this Gal3-ribonucleoprotein complex occurs in the cytoplasmic RNA granules. Like the nuclear bodies, the most prominently studied cytoplasmic RNA granules, P-bodies and stress granules, are membrane-less organelles rich in proteins containing IDR and their assembly is mostly driven by LLPS46.
The CRD of galectin family members is composed of approximately 130 amino acids. As observed in several high-resolution crystal structures, the polypeptide backbone exhibits two β-pleated sheets forming a sandwich-like structure5,47-50 (Fig. 1B). One sheet, designated as the S-face, contains six strands (S1—S6) in which the sugar-binding site resides (strands S4—S6 in particular); the other sheet, the F-face, contains five strands (F1—F5) in which there is a hydrophobic patch for the binding of other ligands (see Section 5). The saccharide-binding area on the S-face can be subdivided into at least four parts (designated A, B, C, D)51, with the defining galactose moiety in site C interacting with a conserved sequence motif of about seven amino acids (Fig. 1A). Interactions involving the other sites with additional monosaccharide units account for higher affinity of binding for larger carbohydrates, such as A hexasaccharide and oligo/polylactosamine, and for the specificities of different galectin CRDs.
The carbohydrate-binding specificities of galectins from mammals, chick, nematode, sponge and mushroom have been extensively studied and reviewed51-53 and is beyond the scope of the present discussion. Suffice it to say that, despite a general similarity in overall folding, the CRDs with distinct amino acid sequences behave differently not only with respect to carbohydrate ligands but also with respect to non-carbohydrate ligands as well. This is strikingly illustrated by the tandem-repeat galectins in which the NH2-terminal CRD (N-CRD) and the COOH-terminal CRD (C-CRD) are usually both sequence and structurally divergent54. In the binding of carbohydrates, for example, the N-CRD of galectin- 8 (Gal8), differs from the CRDs of other galectins, including the C-CRD of Gal8 itself. Gal8 N-CRD binds acidic saccharides (sulfated/sialylated)52,55 such as GM3 and GD1a, oligosaccharides (LacNAc)2 and (LacNAc)552, and the sugar nucleotide NAD55.
Although the C-CRD of Gal8 binds carbohydrate ligands at the S-face, its F-face can bind the autophagy receptor, NDP5255. This observation provided an important advance in terms of one intracellular activity of galectins: Gal8 uses its N-CRD to bind glycans exposed after vesicular rupture caused by pathogens such as Salmonella typhimurium and recruits NDP52 in a selective autophagic response. Although the two CRDs of Gal8 as well as the CRDs of other galectins share a high degree of sequence and structural homology, only the C-CRD of Gal8, thus far, has been reported to bind NDP52. This highly specific interaction is mainly driven by hydrophobic contacts and is supported by hydrogen bonds56,57 but the key selectivity determinant that prevents other galectin CRDs from binding NDP52 appears to be due to steric hindrance at residue 323 (Ala in Gal8 C-CRD; side chains larger than Ala interfere with binding)57. Therefore, the Gal8 C-CRD also appears to be distinctive in terms of binding this particular non-carbohydrate ligand.
Gal8 also binds to TRIM (tripartite motif) proteins (e.g. TRIM5α) as a part of broad interaction between TRIMs and galectins58. Like Gal8, the CRD of Gal3 binds to TRIM proteins, TRIM16 in particular (Table 2), a cargo-receptor-organizer for initiation of autophagy during lysosomal damage58. This binding is dependent on ULK1 serving as a platform. TRIM16, in turn, is connected to mTOR and TFEB, the latter being a transcription factor that coordinates the expression of lysosomal hydrolases and membrane proteins in autophagy. As discussed above, the NTD of Gal3 binds ESCRT components ALIX and Tsg10129,31, participating in processes involved in endolysosomal repair33. The Gal3 CRD, on the other hand, binds to TRIM proteins58, implicated in autophagic removal of damaged lysosomes32.
The Gal3 CRD has a binding site for its own NTD8,15,16 (Table 2); this corresponds to the site on the F-face of the C-CRD of tandem-repeat Gal8 for binding of the autophagic receptor, NDP52,16. As discussed above for the NTD, intermolecular NTD—CRD interactions represent one way by which Gal3 can oligomerize to achieve multivalency18-21. In addition to this scheme of Gal3 oligomerization involving the NTD, there is also self-association mediated by intermolecular homophilic CRD—CRD interactions59-61 (Table 2).
Prototype galectins form dimers through CRD—CRD interactions: (a) Gal1 and galectin-2 (Gal2) form dimers in a side-to-side orientation, with significant contributions by the β-strands F1 and S147-50; (b) galectin-7 (Gal7) forms dimers in the back-to-back orientation, with monomer-monomer contacts between the β-sheets on the F-face50,62; and (c) galectin-5 (Gal5) and galectin-10 (Gal10) form dimers in the face-to-face orientation, with the S-face playing a key role and leading to a much larger interface area and closer distance between the carbohydrate-binding sites of the two monomer units50,54,63. The tandem-repeat galectins Gal864 and galectin-9 (Gal9)65 also exhibit CRD—CRD interactions to form multimers. Although both CRDs are involved in the intermolecular Gal9 interaction, the N-CRD appears to play a dominant role. Moreover, Gal9 also associates with Gal3 and Gal8 but curiously, not Gal165.
Like Gal9’s selective binding to Gal3 and Gal8 but not to Gal1, the cysteine- and histidine-rich protein (Chrp) binds to the CRD of Gal3 (Table 2) but does not bind to the lone CRD in Gal166. Therefore, some ligands appear to be rather selective in terms of which CRD to bind. The Gal3—Chrp binding was not sensitive to the presence of carbohydrate ligands such as lactose or laminin. The details of the selective recognition, such as the participation of S-face or F-face, of the Gal3—Chrp binding, of the CRD—CRD interaction in the self-association of Gal3 and of the CRD—CRD interaction in the binding of Gal9 to Gal8 remain to be elucidated.
The oncogene K-Ras, when loaded with GTP, can bind to the CRD of Gal367 (Table 2). It was proposed that a hydrophobic patch between the two β-sheets of the CRD interacts with farnesylated K-Ras68. This is similar to Gal1 binding to H-Ras69,70, providing a scaffold for the anchorage of Ras nanoclusters to inner surface of plasma membrane. The nanoclusters, in turn, regulate signals derived from growth factor binding to the ERK (Extracellular Signal-Regulated Kinases) branch of the MAP (Mitogen Activated Protein) kinase pathway68,70.
| Liganda | Inhibition by saccharides | Gal3 NTD | No | Gal3 CRD | Yes | CRDs of other galectins | Yes | Tripartite motif proteins (e.g. TRIM16) | Not directly testedb | Cysteine-, histidine-rich protein Chrp | No | Oncogene K-Ras | Not directly testedc | Apoptosis repressor Bcl-2 | Yes | Transcriptional coactivator OCA-B | No | Transcriptional coactivator β-catenin | Yes | General transcription factor TFII-I | Yes | Nuclear transport protein importin-α | Not directly tested | Nuclear transport protein exportin-1 | Not directly tested |
|---|
a NTD, NH2-terminal domain of galectin-3 (Gal3); CRD, Carbohydrate recognition domain.
b TRIM16 colocalized with galectin-3 on Mycobacterium tuberculosis phagosomes with galectin-3 presumably bound to exposed glycoconjugates.
c Lactose had no effect on the corresponding interaction between galectin-1 and H-Ras.
Even when bound with the same ligand, the CRDs of distinct galectins can lead to different physiological outcomes: Gal771, a prototype, and Gal372, a chimera type, each contains one CRD and both will bind to the apoptosis repressor protein Bcl-2. However, Gal7 is pro-apoptotic71,73 and the Gal7—Bcl-2 interaction is not sensitive to lactose inhibition71. In contrast, Gal3 binding to Bcl-2 (Table 2), which can be inhibited by lactose, results in anti-apoptosis72. It is not clear whether Bcl-2 binds to distinct surfaces on the CRD of Gal7 versus that of Gal3, thereby leading to different platforms assembled by the respective CRDs to effect the opposite physiological responses. Since Gal3 and Gal7 are expressed in different cells/tissues and since the studies on the effect of Bcl-2 binding to the CRDs of Gal3 and Gal7 were investigated in distinct cell types, an alternative possibility is that the apoptotic pathways affected may be cell type-specific. The tandem-repeat galectin-12 (Gal12) also promotes apoptosis, although a direct intracellular interaction with Bcl-2 has not been reported74.
In nuclear extracts of HeLa cells, the cell-specific transcriptional coactivator OCA-B will bind the CRDs of Gal1, Gal3, and Gal875 and the general transcription factor TFII-I will bind the CRDs of either Gal1 or Gal376. However, while the OCA-B—CRD interaction is not affected by saccharide ligands, the binding of TFII-I to the CRDs of Gal1 and Gal3 is sensitive to saccharide inhibition. The binding of TFII-I to Gal1 and Gal3 was of particular interest because identification of the former as a component of purified spliceosomes77 and the demonstration of the latter as required factors in cell-free splicing assays35,38. Hirabayashi and Kasai had shown that substitution at Asn 46 in the CRD of Gal1 resulted in the loss of its saccharide-binding activity78. Although the site-directed mutant N46D of Gal1 showed drastic reduction of carbohydrate-binding capacity, it retained the ability to bind TFII-I, as well as the ability to reconstitute pre-mRNA splicing in a galectin-depleted nuclear extract76. These results suggest that saccharide-binding, per se, was not necessary for the interaction of Gal1 with the nuclear splicing machinery. Nevertheless, saccharides that bind to Gal3 with high affinity inhibit the Gal3—TFII-I interaction76 and the cell-free pre-mRNA splicing assay38,79. This is probably best explained by the hypothesis that carbohydrate binding to the saccharide-binding site induces a conformational change that affects the interface of the CRD with non-carbohydrate ligands.
The CRD of Gal3 also binds β-catenin, a co-activator of the Tcf/Lef (T-cell factor/lymphoid enhancer factor) transcription factor family in the Wnt signaling pathway80 (Table 2). This binding is sensitive to inhibition by saccharide ligands such as lactose. In this connection, it may also be interesting to note that nuclear localization of the protein Dishevelled (Dsh) is required to serve as an intracellular mediator of signals activated by Frizzled receptors to the Wnt/β-catenin pathway81. Curiously, both Dsh and Gal3 share similar motifs in terms of their nuclear localization signal (NLS) and nuclear export signal (NES): (a) the NLS is of the “IXLT” motif81,82; and (b) the NES is of the “leucine-rich” motif with requisite spacing of hydrophobic Leu and Ile residues and is recognized by CRIM1 exportin81,83 (Table 2). In murine Gal3, the NLS is 253ITLT256 at the top of strand F1 of the F-face and the NES begins with 241L in the sole α-helix and ends with 249I buried strand S2 of a β-sheet (Fig. 1B). In human Gal3, the NLS (223HRVKKL228) binds to the nuclear import receptor importin-α84 (Table 2). Thus, the CRD of Gal3 has sequences critical in binding to the transport receptors mediating its shuttling between the nucleus and cytoplasm85.
Dimer formation by the prototype galectins Gal1 and Gal2 occurs through specific interactions of β-strands on the F-face and S-face of one subunit with the corresponding sites on the other subunit47-50. Similarly, the Gal7 dimer arises through monomer-monomer contacts between the β-sheets on the F-face50,62 and the Gal5 and Gal10 dimers are based on contacts between the S-faces50,54,63. None of these interactions are sensitive to the simultaneous presence of galectin saccharide ligands. On the other hand, tandem-repeat Gal864 and Gal965 can also form dimers and the N-CRD of Gal9 can bind to the CRDs of Gal8. In contrast to the prototype galectins, these CRD—CRD interactions are inhibited by lactose. Similarly, the lone CRD in the chimera-type Gal3 can mediate the protein’s intermolecular homophilic association that is sensitive to saccharide inhibition59,60. So, what is the structural basis that makes these latter CRD—CRD interactions distinct from those involved in the dimerization of the prototype galectins? In this connection, Miyanishi et al.65 also showed that while the Gal9 CRDs can interact with the CRDs of tandem-repeat Gal8 and chimera-type Gal3, Gal9 failed to bind to prototype Gal1. Therefore, a corollary question might be: does the dimerization scheme of prototype galectins preclude further CRD—CRD interactions, either in formation of higher homophilic oligomers or in binding to the CRDs of other galectins? Indeed, the recent data of Eckhardt et al. suggest that homodimerization of Gal1 weakens the interaction of Gal1 with the extracellular ligand chemokine CXCL12 to form heterodimers86.
The intrinsically disordered NTD of Gal3 can interact with its own CRD8,15,16. In the same vein, could interactions between Gal3 NTD and the C-CRD of Gal9 account for, at least in part, the observed binding of Gal9 to Gal365? AlphaFold is a neural network-based computational method for predicting protein structures87. In a recent assessment of AlphaFold2 applications, Akdel et al.88 documented that polypeptide regions with low-confidence predictions in AlphaFold2 are enriched for IDRs. Bcl-2 (e.g. residues 30—75), OCA-B (e.g. residues 1—25), TFII-I (e.g. residues 240—340, 680—710, 960—990), and β-catenin (e.g. residues 30—55, 710—780) are all included in their Supplementary Dataset on IDRs88. Could the IDRs of Bcl-2, OCA-B, TFII-I, and β-catenin interact with the Gal3 CRD (Table 2), much like the NTD of Gal3 interacting with its own CRD? For example, Yu et al. showed that residues 7—43 at the NH2-terminus of OCA-B were sufficient for galectin binding75. In this connection, it should also be noted that Eckhardt et al. suggest that a structured region rather than an IDR can compete against the IDR in the Gal3 NTD in interacting with CRD86.
The binding of some non-carbohydrate ligands to CRDs can be inhibited by saccharide ligands (e.g. Gal3—Bcl-272; Gal1/Gal3—TFII-I76) but the binding of other non-carbohydrate ligands is insensitive to saccharide ligands (Gal3—Chrp66; Gal7—Bcl-271; Gal1/Gal3—OCA-B75). What, for example, distinguishes the interaction of Bcl-2 with the CRD of Gal3, sensitive to lactose inhibition and leading to anti-apoptosis, with the corresponding interaction of Bcl-2 with the CRD of Gal7, insensitive to lactose and pro-apoptotic? Does Bcl-2 bind to distinct sites on the CRD of Gal3 versus that on the CRD of Gal7 because the latter dimerizes through CRD—CRD interactions? Neither the CRD—NDP52 interaction55 nor the galectin CRD—TRIM proteins interaction58 is sensitive to the simultaneous presence of carbohydrate ligands inasmuch as their biological activities serve the purpose of sensing glycoconjugates on damaged intracellular vesicles that lead to the recruitment of proteins involved in the autophagic response.
The carbohydrate binding ability of Gal351-53, the Gal3 NTD—ESCRT interaction29,31 and the Gal3 CRD—TRIM interaction58 all serve to bridge two inter-connected processes: recognition of exposed glycans from damaged endolysosomal membranes and repair or autophagic removal of damaged lysosomes32. Similarly, the Gal3 NTD—hnRNP/snRNP interaction34,39 and the Gal1/3 CRD—TFII-I interaction76 connect the processes of RNA biogenesis and maturation. The NTD of Gal3 is an IDR (Fig. 1B), whose special feature is conformational heterogeneity allowing it to bind multiple ligands14. Are there other inter-connected processes that can be bridged by this chimera-type galectin? In addition to binding carbohydrates, the CRDs of various galectins can also bind a myriad of non-carbohydrate ligands (as illustrated in Tables 1 and 2 for Gal3). Are there other examples of galectins, particularly the tandem-repeat galectins whose N-CRD and C-CRD behave distinctly, that serve to bridge inter-connected processes through interaction with non-carbohydrate ligands?
I thank Drs. Jun Hirabayashi and Sachiko Sato for critically reading the manuscript and for their helpful suggestions in revision. In particular, Dr. Sato indicated that some non-carbohydrate ligands such as Bcl-2, TFII-I, and β-catenin contain intrinsically disordered regions in their respective polypeptides and raised the possibility that these regions provided the basis for their observed interaction with the carbohydrate recognition domain of galectin-3. The work carried out in the author’s laboratory has been supported by grant GM-38740 from the National Institutes of Health and by the Michigan AgBioResearch Project MICL02455.