
Yukiko Yoshida
Laboratory Head
Laboratory of Protein Metabolism, Tokyo Metropolitan Institute of Medical Science
She received her Ph.D. in1994 from the Graduate School of Agricultural and Life Sciences at the University of Tokyo, and then worked as a postdoctoral fellow studying glycobiology at RIKEN (Wako). She has been working at the Tokyo Metropolitan Institute of Medical Science since 1997. Her research focus has been on the physiological functions of the ubiquitin system-related glycoproteins.
The ubiquitin-proteasome system is the major pathway for selective degradation in eukaryotes. Ubiquitin attaches to lysine residues of a substrate, thereby forming ubiquitin chains with various topologies and linkages. Distinct ubiquitin chains serve diverse functions. Recent reports have emerged on the ubiquitination of non-proteinaceous biomolecules, such as lipids and sugars, signifying a rapid expansion of the field of ubiquitination. Here, I introduce the sugar-mediated ubiquitination of Nrf1, a transcription factor of the proteasome, which occurs in peptide: N-glycanase (NGLY1) deficiency, a rare disease.
Ubiquitination is mediated by a cascade of three distinct kinds of enzymes: 2 kinds of ubiquitin-activating enzymes (E1s), ~ 30 kinds of ubiquitin-conjugating enzymes (E2), and more than 600 kinds of ubiquitin ligases (E3). Ubiquitination generally involves the formation of isopeptide bonds between the C-terminal glycine of ubiquitin and the ɛ-amino group of the lysine side chains of substrate proteins1-3.
The attached ubiquitin can be further ubiquitinated on its own lysine residues (K6, K11, K27, K29, K33, K48, K63) or the α-amino group of the N-terminal methionine to form polyubiquitin chains. Among them, K48- and K63-linked ubiquitin chains are the most abundant and together account for 70-90% of all linkage types in mammals. K48-linked chains serve as signals for proteasomal degradation, and K63-linked chains act as signals for endocytosis, DNA damage responses, and intracellular signaling events4-6.
The removal of ubiquitin chains is catalyzed by deubiquitinating enzymes (DUBs) following the completion of their respective functions. Consequently, the ubiquitination reaction is a reversible and dynamic process (Fig. 1)5.
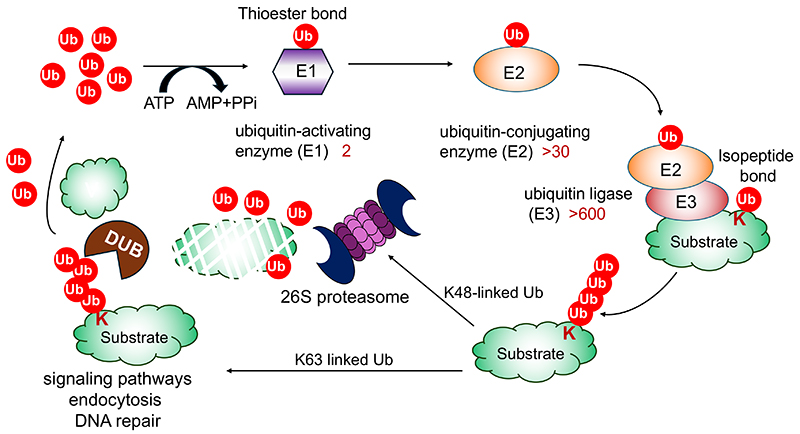
Lysine residues are the canonical ubiquitination sites, given the fact that more than 60,000 lysine residues modified with ubiquitin are annotated in the proteome8. However, ubiquitin can be attached to initial methionine (peptide bond), serine or threonine (oxyester bond), or cysteine (thioester bond) residues in proteins9-12.
Furthermore, non-proteinaceous substrates including the lipid A component of lipopolysaccharides (LPS) of Salmonella, phosphatidylethanolamine (PE), ADP-ribose, and glycogen can be ubiquitinated13-17. Although the significance of non-canonical ubiquitination events remains to be fully elucidated, their occurrence is more widespread than currently supposed.
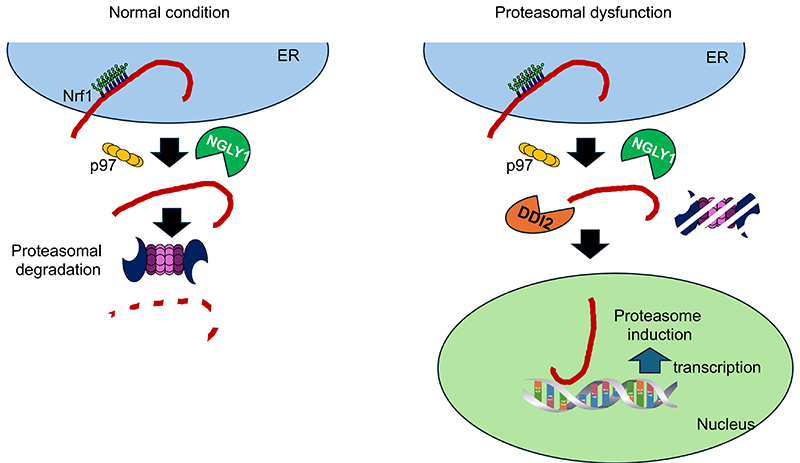
The ubiquitin-proteasome system is essential for cell survival, as it regulates cellular events including the cell cycle and DNA repair18. The proteasome is a large protein complex composed of 66 subunits, and its expression is regulated by the transcription factor Nrf119,20. Nrf1 is synthesized as an N-glycosylated, single-pass membrane protein in the endoplasmic reticulum (ER). In general, misfolded proteins in the ER are retrotranslocated to the cytosol by p97, an AAA-ATPase complex, and degraded by proteasomes, a process called “ER-associated degradation” (ERAD) (Fig. 2). At least under steady state conditions, Nrf1 is constitutively targeted to the ERAD pathway, probably due to its large unstructured domain in the ER, and the amount of the Nrf1 is maintained at low levels in the cells21,22. Under conditions of compromised proteasomal activity, Nrf1 escapes degradation and is transported to the nucleus, where it induces the expression of genes encoding proteasome subunits. In this process, Nrf1 activation requires cleavage by DNA-damage inducible 1 homolog 2 (DDI2) and deglycosylation by peptide: N-glycanase (NGLY1)23-26. Mutations in NGLY1 gene result in NGLY1 deficiency, a rare autosomal recessive congenital disorder, and the expression of the proteasome is reduced in this disease27.
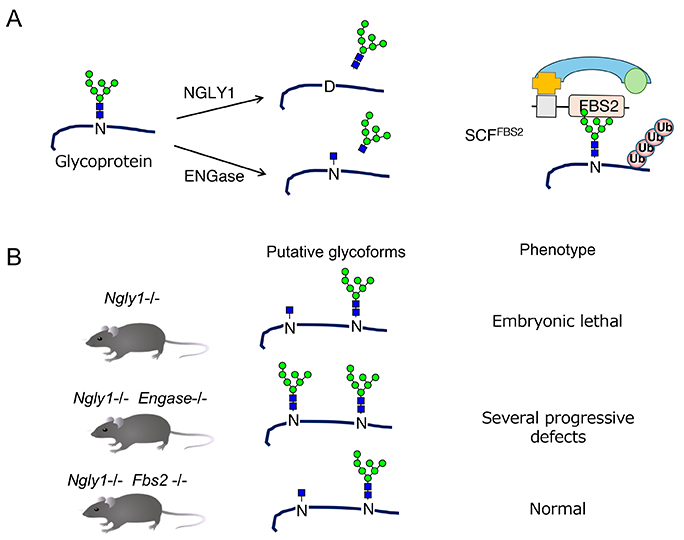
As animal models for NGLY1 deficiency, NGLY1 orthologs-deficient models such as C. elegans, Drosophila, mice, and rats, have provided important findings for NGLY1 study (28-30). Notably, the knockout of NGLY1 in Ngly1-KO mice is embryonic lethal on a C57BL/6 genetic background, indicating that NGLY1 activity is essential for their development. Interestingly, the absence of a gene encoding endo-β-N-acetylglucosaminidase (ENGASE), another cytosolic deglycosylating enzyme, partially rescues Ngly1-KO mice from embryonic lethality, yet the double-KO mice show some progressive defects (Fig. 3)31. We observed that additional deletion of the FBS2 gene completely restored the viability of Ngly1-KO mice, and that Ngly1;Fbs2-dKO mice grow without abnormalities. FBS2 is a component of the SKP1-CUL1-F-box (SCF) E3 ubiquitin ligase complex and recognizes N-glycans of glycoproteins32. Therefore, FBS2 ubiquitination activity seems to be the primary cause of the lethality in Ngly1-KO mice. In addition, FBS2 overexpression in NGLY1-KO cells induces cell death, indicating ubiquitination of certain glycoproteins is detrimental to cells lacking NGLY1 activity33.
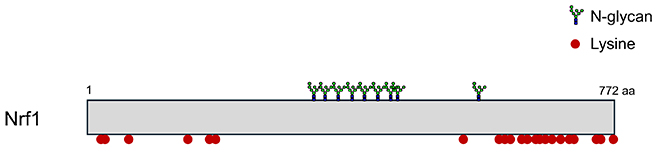
We found that Nrf1 was highly ubiquitinated in FBS2-overexpressing NGLY1-KO cells. However, lysine residues, the usual ubiquitination sites, are not found in Nrf1 around the sites of N-glycosylation recognized by FBS2 (Fig. 4). Furthermore, Nrf1 ubiquitination was not detected in NGLY1:ENGASE-dKO cells, but ubiquitination was restored by expressing ENGASE, indicating that Nrf1 ubiquitination is required for FBS2 and ENGASE activities in the absence of NGLY134. Since ENGASE generates N-GlcNAc on glycoproteins, we hypothesized that SKP1-Cullin1-F-box protein-RBX1 (SCFFBS2) E3 ubiquitin ligase complex could ubiquitinate N-GlcNAc on Nrf1 (Fig. 3A)34.
Using an in vitro ubiquitination reconstitution system with chemically synthesized glycopeptides, we found several unique features of this ubiquitination as follows (Fig. 5). First, this ubiquitination occurs through oxyester bonds on serine or threonine residues near N-glycans, as well as the 6-position hydroxy group of N-GlcNAc residues generated from deglycosylated by ENGASE. Second, SCFFBS2 cooperates with another E3 ligase ARIH1. ARIH1 is known to bind to neddylated Cullin-RING E3 ligases (CRLs) and mediate the addition of the first ubiquitin on substrate35,36, but in this case SCFFBS2 captures N-glycans of Nrf1, and ARIH1 ubiquitinates them. Third, the ubiquitin chains are complex, branched, and attached through K6, K11, K33, and K48 ubiquitin linkages. As a result, the atypical ubiquitin chains inactivate Nrf1 by preventing its nuclear transport34.
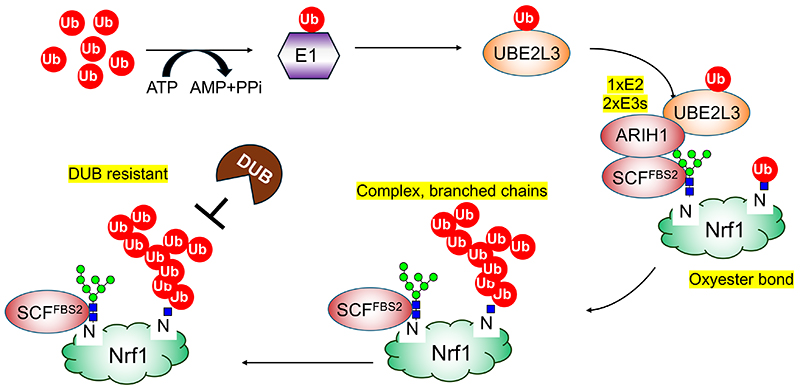
I introduced here that ubiquitination also occurs in the glycans of glycoproteins. While the enzymes involved in ubiquitination are located in the cytoplasm and nucleus, the sugar chains are usually located outside the cell or inside the secretory organelles (endoplasmic reticulum, Golgi apparatus, lysosomes, endosomes), and the two are separated by a membrane. Therefore, for a glycoprotein to be ubiquitinated, it must appear in the cytoplasm or in the nucleus (37). In the case of Nrf1, it is thought that this appearance in the cytosol is due to ER-associated degradation, but there are several other possibilities, such as damage to organelles by bacteria or foreign cells that have invaded the body. It will be interesting to see whether such ubiquitination of glycoprotein sugars is seen in other proteins, and whether more non-proteinaceous ubiquitination will be discovered in the future.