
Kiyoko F. Aoki-Kinoshita
Deputy Director, Glycan Life System Integration Center (GaLSIC) at Soka University
She graduated from Northwestern University, USA and received her Ph.D. in 1999. She worked as a postdoctoral fellow at Academia Sinica, Taiwan (2000) and the Institute for Chemical Research, Kyoto University (2003). She then has worked at Soka University since 2006 and became Professor in 2014 (Bioinformatics department), appointed as Deputy Director at GaLSIC in 2021.

Yusuke Matsui
2014 - Completed Ph.D. program in Information Science at Hokkaido University, Japan, finishing 1 year and 9 months ahead of schedule.
2015 - Designated Assistant Professor at the Center for Neurological Diseases and Cancer, Graduate School of Medicine, Nagoya University / Leader of the Real World Data Circulation Leaders, Program for Leading Graduate School Nagoya University.
2018 - Associate Professor at the Biomedical and Health Informatics Unit (Principal Investigator), Department of Integrated Health Science, Graduate School of Medicine, Nagoya University.
2022 - Head of the Systems Biology Division at the Integrated Glycan-Big Data Center (iGDATA), Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System.

Yukie Akune-Taylor
Project lecturer, Glycan Life System Integration Center (GaLSIC) at Soka University
She graduated from Soka University and received her Ph.D. in 2016. She then worked as a postdoctoral fellow at Glycosciences Lab, Imperial College London (UK). She is also completed her Ph.D. in molecular biology (Macquarie University, Australia) in 2017. She has been in her current position since 2023.

Achille Zappa
Project lecturer, Glycan Life System Integration Center (GaLSIC) at Soka University, Hachioji, Japan.
He achieved the Master Degree in Biomedical Engineering, at Università degli Studi Genova, Italy. He attained a Ph.D. in Bioengineering from School of BioEngineering at Università degli Studi di Genova (DIBRIS), Genoa and IRCCS AOU San Martino Uni Hospital - IST National Cancer Research Institute, Italy. He then worked as a postdoctoral fellow at University of Galway – DSI and Insight Data Research Center in Galway, Ireland. He has been in his current position since 2024.
The Human Glycome Atlas (HGA) project is a collaborative effort between the Institute for Glyco-core Research (iGCORE) at Nagoya University, the Exploratory Research Center on Life and Living Systems (ExCELLS) at the National Institutes of Natural Sciences, and the Glycan and Life Systems Integration Center (GaLSIC) at Soka University. Our aim to elucidate the glycans and biosynthetic mechanisms in humans. This paper provides an overview of the Knowledgebase TOHSA being developed by HGA Segment 4 (TOHSA Development Unit).
Various experimental techniques such as mass spectrometry, liquid chromatography, glycan microarrays and lectin microarrays have been developed to analyze glycan structures and functions, glycan-glycan binding sample interactions and other correlations. Glyco-informatics research includes the development of technologies to accumulate, systematize and compile data resources based on glycan-related experimental data, as well as to analyze glycan structures and their functions. This includes analyzing glycan binding and sample correlation using molecular dynamics, including the incorporation of biological and/or biochemical information. Currently, many open-source glycan-related databases and repositories are being published. For example, GlyTouCan1 is the first and only repository where users can register glycan structures. A registered glycan structure in GlyTouCan is assigned a unique accession number, which can be used as a reference in papers and linked to other databases. Over 240,000 glycan structures are registered in GlyTouCan at the time of this writing, and the number of registrations is increasing every year. Another example of a glycan-related data resource is GlyCosmos2,3. GlyCosmos is a web portal that provides a range of information on glycan structures, glycan-related genes, glycoproteins, glycolipids, reaction pathways and disease-related glycans regardless of species.
Most databases currently available are based on the secondary use of data, where data are collected and organized according to their purpose. Users can use these databases to efficiently search for their target data and related information. Moreover, bioinformaticians can develop other databases or data analysis tools for the tertiary use of data. On the other hand, during the data collection it is sometimes necessary to liaise with the researchers who reported the data; this may involve tasks such as reformatting the data for entry into a database and verifying the version of the data. Additionally, until now, no one has developed a comprehensive glycan-related resource for human; there is no standardized Reference Human Glycome by which researchers can benchmark human glycomics (including glycoconjugate) data. One of the goals of the HGA project4 is to build Knowledgebase TOHSA (Total Human Saccharide Atlas)5. HGA Segment 4, under the leadership of Kiyoko F Aoki-Kinoshita (Soka University), is responsible for the development and management of TOHSA4. Segment 4 consists of (i) the Integrated Analysis Infrastructure Unit and (ii) the TOHSA Development Unit. The Integrated Analysis Infrastructure Unit, led by Yusuke Matsui (Tokai National Higher Education and Research System), develops large-scale information systems, analysis pipelines and tools, and analytical results6. The TOHSA Development Unit designs, develops and manages comprehensive databases and interfaces that are user-friendly and incorporate Semantic Web7 and AI Technologies. A semantic knowledge base is a resource that not only aggregates data stored in a database but also integrates and contextualizes 'knowledge' related to that data. This knowledge is also derived by integrating and connecting data from other multiple public data resources, enabling more sophisticated data relationships and insights. The Knowledgebase TOHSA stores experimental data, large cohort data (primary use) and analysis tools generated in the HGA project, together with related data collected using Semantic Web technologies. The database will initially be available to users on a limited access basis, after which the published data will be open access (secondary use).
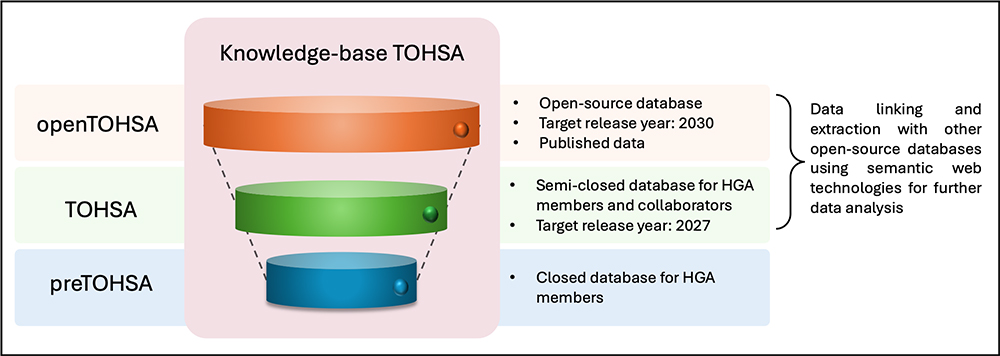
Data integration into Knowledgebase TOHSA consists of three main stages (Figure 1). The first stage is the deposition stage whereby experimental data and related metadata, generated internally within the HGA project, are stored in a repository called “preTOHSA.” The data generated by researchers involved in this project need to be uploaded to the repository, analyzed and feedback given without delay. The key role of preTOHSA at this stage is to serve as the foundation for the data infrastructure and to store the data as big data in a structured manner. This will allow us to expand seamlessly into big data analysis. The definition and management of metadata is an important element to ensure the quality of the data, especially when considering the use of the data for future analysis (tertiary use). The metadata will store information in accordance with relevant guidelines. For example, metadata associated with experiments such as sample preparation and mass spectrometry will be stored in accordance with the Minimum Information Required for A Glycomics Experiment (MIRAGE)8 recommended by the MIRAGE Commission which consists of experts in the respective fields of glycoscience. The sample preparation guidelines primarily suggests the recording of information about from what the sample is derived and how it was isolated, chemically modified, and purified9. The mass spectrometry guidelines recommend descriptions of the type of experimental equipment and control parameters related to the experimental equipment, as well as data acquisition protocols10.
The second stage is the main database, TOHSA, which will contain data migrated from those datasets stored in preTOHSA that are restructured and ready to be shared with collaborators. TOHSA is planned to be released (via limited access) by the fifth year of the HGA project. The data in TOHSA will not only include comprehensive analyses and cataloging of glycome and glycoproteome information from qualitative and quantitative assessments of blood samples from several large cohorts, such as the Dementia Cohort and the Healthy Elderly Cohort, but it will also store other additional glycan-related information. This information will be meticulously integrated, structured and standardized using advanced Semantic Web technologies.
The third stage will involve the development and management of “OpenTOHSA” which will be an open-access knowledgebase portal based on the data published through the HGA project and will be released near the end of the project. OpenTOHSA is expected to be widely used not only by glycobiologists but also by all scientists involved in life sciences, medical research, and students wishing to join the field of life science.
The experimental data generated by the HGA project are divided into three main categories, which correspond to the three Segments of the HGA project. These are: Glycoproteomic data covering all human glycoproteins (Segment 1), individual glycoproteomic and glycomic analysis data from large cohorts (Segment 2), and a glycan biosynthesis atlas for glycan biosynthesis mechanism analysis (Segment 3). Raw data generated from mass spectrometry, enzyme chemistry and imaging will be stored in the knowledgebase TOHSA along with associated experimental metadata and processed datasets.
The data stored in the knowledgebase TOHSA will then be further analyzed and its results supplemented by links to external databases such as genomics and proteomics databases. At the end of the project (in eight years), we plan to create a standard for all relevant human glycan information. This will be linked to other glycan information through GlyCosmos and the GlySpace Alliance11, an international organization whose one goal is to provide an informatics framework for glycan-related multi-omics data to the community.
Knowledgebase TOHSA aims to build a comprehensive structured and annotated resource based on Semantic Web technologies including cataloged information from clinical data and analysis results of tens of thousands of samples obtained through this project. It is also crucial to develop user-friendly interfaces and visualization tools that are intuitive and accessible. These tools should be designed to meet the needs of biologists, students, and other users and use-cases, ensuring they can easily navigate and utilize the data effectively. In the future, we intend to develop web tools that incorporate semantic technologies and AI models that can be used for predictive analyses such as pathway prediction and glycan-protein binding. By providing an integrated and standardized semantic knowledgebase of glycome information, the HGA project and TOHSA aim to accelerate glycoscience research and potentially lead to breakthroughs in the elucidation of disease mechanisms and the development of novel therapeutic approaches.