
Hisatoshi Hanamatsu
Specially Appointed Lecture
Institute for Glyco-core Research, Nagoya University, Tokai National Higher Education and Research System
He received his PhD in life sciences from Hokkaido University in 2014. He did postdoctoral training at Hokkaido University from 2014 in Graduate School of Advanced Life Science and Frontier Research Center for Post-Genome Science and Technology, Hokkaido University. He did postdoctoral training at Hokkaido University from 2017 in Department of Advanced Clinical Glycobiology, Faculty of Medicine and Graduate School of Medicine, Hokkaido University. In 2020, he was a Specially Appointed Assistant Professor in Faculty of Medicine and Graduate School of Medicine, Hokkaido University. Form 2023, he is a Specially Appointed Lecture in Institute for Glyco-core Research, Nagoya University. His research interests focus on the development of sialic acid linkage-specific derivatization and total glycome analysis.

Masaki Kurogochi
Research Scientist
Institute for Glyco-core Research, Nagoya University, Tokai National Higher Education and Research System
Masaki Kurogochi graduated from Hokkaido University in 1998, got a doctoral degree in science. He worked as a senior researcher in Northern Advancement Center for Science and Technology(2004), a postdoctoral researcher in Hokkaido University (2005), became an assistant professor at the Faculty of Advanced Life Science, Hokkaido University. In 2012, he joined The Noguchi Institute, where he worked as a research scientist in Laboratory of Glycobiology, a leader in Laboratory of Glyco-bioengineering, a research scientist in Laboratory of Glyco-organic Chemistry. He has been in his current position since 2024. His research interests are focused on organic chemistry, analytical chemistry, glycoengineering, glycomics and glycoproteomics.

Jun-ichi Furukawa
Designated Professor
Institute for Glyco-core Research, Nagoya University, Tokai National Higher Education and Research System
Visiting Professor
Department of Orthopaedic Surgery, Faculty of Medicine and Graduate School of Medicine, Hokkaido University
Jun-ichi Furukawa graduated from Faculty of Science at Hokkaido University in 1996, received his Ph.D. in Environmental Earth Science at Hokkaido University in 2001. Then, he worked as a JSPS postdoctoral, a postdoctoral fellow (2002–2006), and as a research assistant professor (2006–2016) in Faculty of Science, Hokkaido University. In 2016, he was a Specially Appointed Associate Professor of Faculty of Medicine, Hokkaido University. He has been in his current position since 2022. His current research interest include bioorganic chemistry and analytical chemistry focused on glycans. He is studying the comprehensive glycomic analysis using his original analytical tools.
Carbohydrate chains (glycans) are called the third life chain following nucleic acids (genes) and proteins. A large variety of glycans (sub-glycans), which are linked to proteins and lipids to form glycoconjugates, coat the cell surface and play important roles in biological events. Structural analysis of glycans is essential to understand the functions of glycans and glycoconjugates. The proteome is the entire set of proteins produced or modified by an organism or system, and proteomics is the large-scale study of entire proteomes. Similarly, the glycome is the entire set of glycans produced by an individual organism, and glycomics is their complete analyses. Most glycans are present as a variety of glycoconjugates such as glycoproteins, glycosphingolipids, proteoglycans, and glycosylphosphatidylinositol (GPI) anchors. To elucidate their functions, it is necessary to measure the expression of glycans corresponding to various classes of glycoconjugates. In general, however, alteration of one class of glycan is measured using a transgenic or knockout mouse in which a glycosyltransferase gene is targeted. In fact, the relationship between different sub-glycans has been scarcely analyzed. It has often been reported that there is no clear phenotype in transgenic or knockout mice in which various glycosyltransferase genes are targeted1. This result suggests that comprehensive glycomic analysis of glycoconjugates is needed to elucidate their functions.
Previously, we reported a series of methodologies to prepare and analyze N-glycans, O-glycans, glycosaminoglycans, glycosphingolipid-glycans, and free oligosaccharides by mass spectrometry (MS) and high-performance liquid chromatography2,3,4,5. Each procedure to analyze different classes of glycoconjugate glycans was combined and the entire sub-glycan components could be visualized in the cellular or serum glycome, the so-called total glycome6,7. Recently, we have developed and improved the technologies for glycomic analyses and established sialic acid linkage-specific alkylamidation (SALSA) to distinguish sialylated glycan isomers by MS analysis8.
In segment 2 of the Human Glycome Atlas Project (HGA), the generation of a large-scale human glycome catalog (total human plasma glycome) is one of the most important missions to construct a database called TOHSA. In this section, we introduce our latest studies, covering 2) the improved analysis of N-glycans by a glycoblotting method in combination with sialic linkage-specific derivatization to distinguish sialylated glycan isomers, and 3) the development of an automated N-glycan preparation instrument for large-scale glycomic analysis of human plasma/serum.
N-glycosylation is one of the most common post-translational modifications of proteins, and N-glycans are directly attached to asparagine residues. N-glycans are classified into four types, namely, paucimannose-, oligomannose-, hybrid-, and complex-type N-glycans, according to their unique structures. In human serum, more than 50% of secreted proteins were reported to be glycosylated. N-glycans are readily released from proteins by a very versatile enzyme called peptide-N-glycosidase F (PNGase F). Furthermore, reduction, alkylation, and trypsin digestion of glycoproteins increase the cleavage efficiency of N-glycans by PNGase F9. This reaction mixture contains large amounts of contaminants such as trypsin-digested peptides. Therefore, released N-glycans must be purified for glycomic analysis. We have purified released glycans by a glycoblotting procedure via chemoselective ligation2. Released glycans have a hemiacetal group (aldehyde group) at the reducing end, and a few other compounds have this functional group in human serum. Therefore, glycans are selectively captured by a glycoblotting method.
Most N-glycans have sialic acid residues at the non-reducing end, and their glycosidic linkages are labile due to the α-glycoside bond, making it difficult to quantitatively measure sialylated glycans by MALDI-TOF MS10. In the glycoblotting procedure, sialylated glycans can be derivatized on the solid phase. This modification method of sialic acid residues is introduced in the next section in detail. Finally, N-glycans are released from BlotGlyco beads and simultaneously labeled for MS analysis. The streamlined protocol of N-glycan preparation for MS analysis is shown in Figure 1. The detailed experimental protocol of the glycoblotting method has been described previously3.
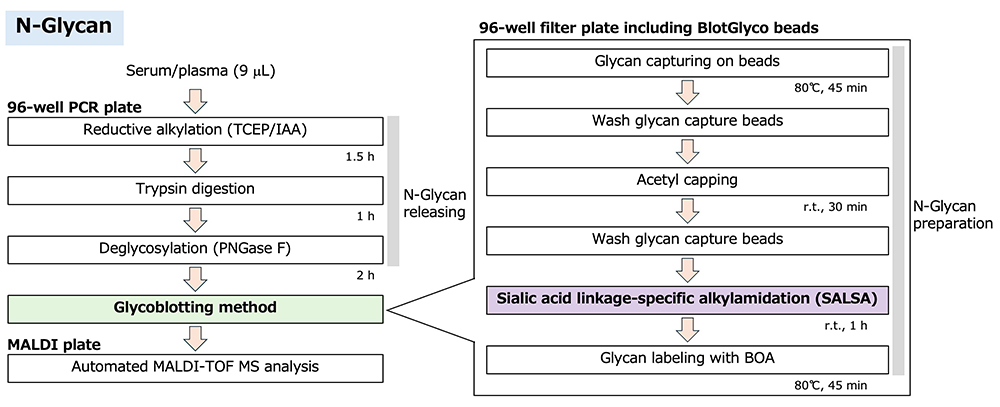
Sialic acid is an acidic monosaccharide present at the non-reducing ends of carbohydrate chains via α2,3-, α2,6-, and α2,8-linkages, and these different linkages are associated with carbohydrate-protein interactions, cellular recognition, and cell signaling11. The α-glycosidic linkages of sialic acid are labile, resulting in loss of terminal sialic acid residues of glycans during MS analysis. In addition, structural isomers with different linkages cannot be distinguished by MS analysis because they have the same molecular weight. In recent studies, several unique sialic acid linkage-specific derivatizations have been reported to distinguish glycan isomers by MS analysis12. We have also developed a linkage-specific derivatization. The strategy of linkage-specific derivatizations is shown in Figure 2.
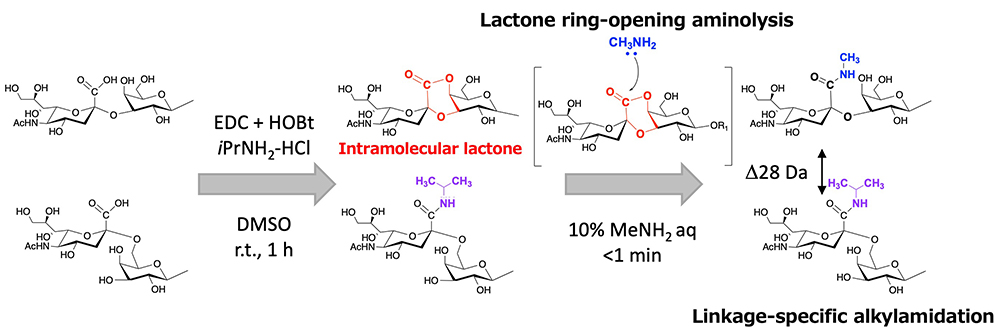
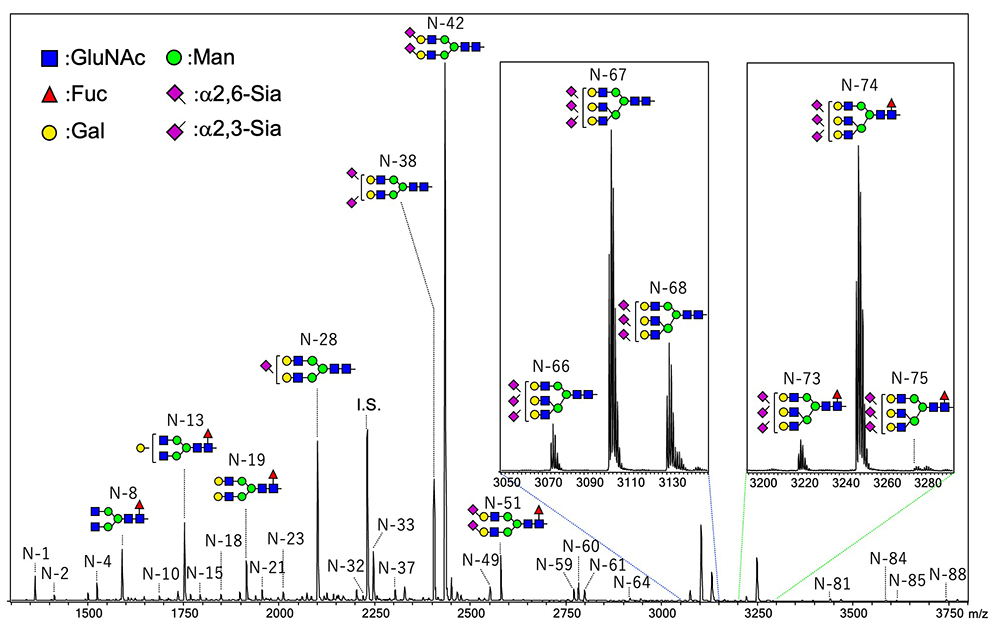
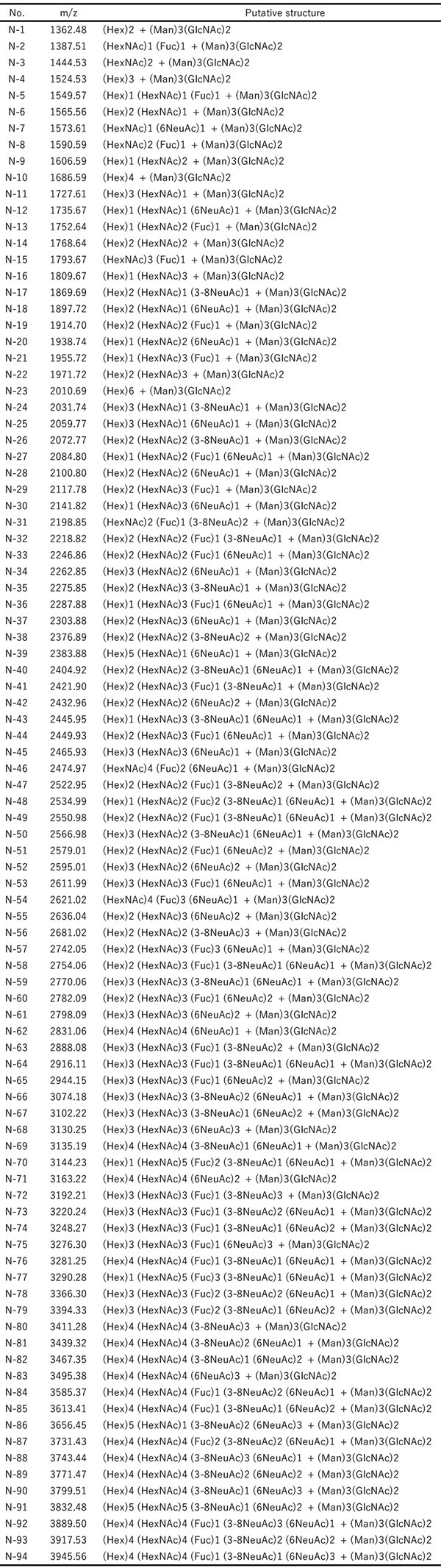
In our procedure, N-glycans containing sialic acids are captured on the solid phase by the glycoblotting method. The first step of condensation can proceed on the solid phase, and carboxylic acid of α2,6-linked sialic acid is converted to iso-propylamide, whereas that of α2,3- and α2,8-linked sialic acid forms intramolecular lactone. We developed a reaction for specific amidation of α2,3- and α2,8-linked sialic acid via intramolecular lactone. This reaction is a very simple process that only requires addition of amine solution to a 96-well filter plate including glycan captured beads, which allows lactone ring-opening and aminolysis. Thus, the reaction time for the SALSA method is reduced from 2 h to 1 h. MALDI-TOF MS spectra of N-glycans in human serum are shown in Figure 3. By the SALSA method, triantennary tri-sialyl N-glycan (A3) is observed as three signals of structural isomers (A33,3,6 m/z 3074.19, A33,6,6 m/z 3102.24, and A36,6,6 m/z 3230.27). Interestingly, the two signals of their fucosylated glycans (A3F) are observed at m/z 3248.30 (A3F3,6,6) and m/z 3221.24 (A3F3,3,6) with a similar ratio, while A3F with all α2,6 linkages (A3F6,6,6) were hardly detected in comparision with A36,6,6.

As described above, we are ready to analyze N-glycans in human serum/plasma. We have developed an automatic glycan preparation instrument for N-glycan analysis in a large-scale cohort study. This instrument is shown in Figure 4. For a 96-well plate format, eight-channel multi-syringes are equipped. The thermostatic oven can be controlled at temperatures ranging from room temperature to 100°C. Denatured serum/plasma proteins are digested with trypsin and PNGase F in a 96-well PCR plate at 37°C. Released N-glycans are also trapped and labeled on a 96-well filter plate including BlotGlyco beads in a thermostatic unit. The SALSA reaction, which requires vigorously mixing, can be performed on a 96-well plate shaker. Finally, purified and derivatized N-glycans in serum/plasma are prepared and spotted on a MALDI plate. Using this automatic instrument, all processes for N-glycan preparation from 96 serum/plasma samples can be completed within 16 h.
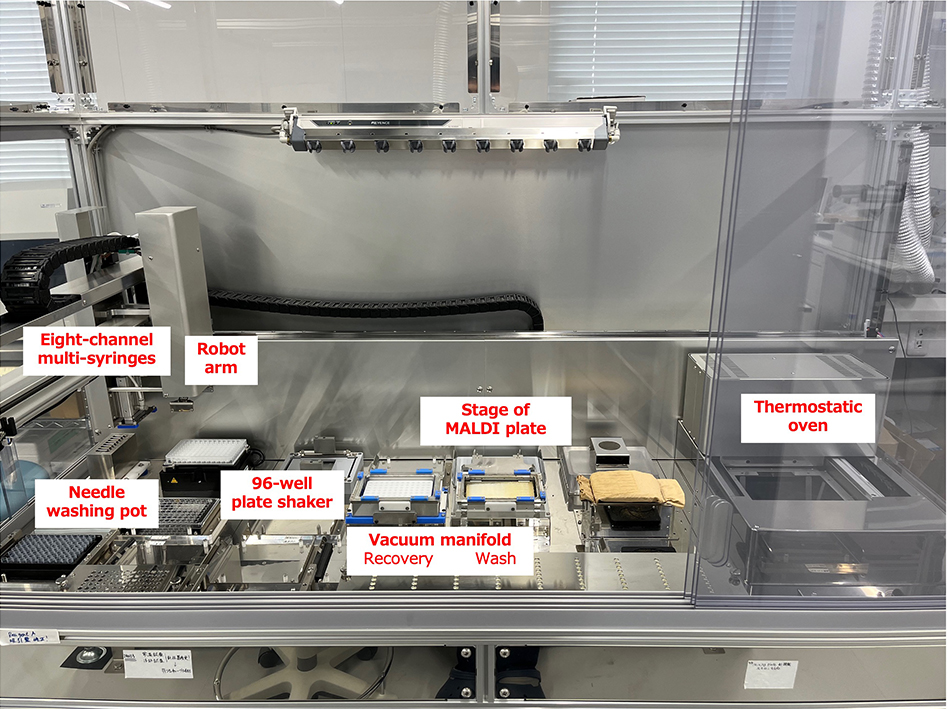
We are ready to measure N-glycans in serum/plasma in large-scale cohort studies, and a maximum of 96 samples/day can be automatically measured. The development of glycomics tools to analyze the enormous MS spectra is one of the most important hurdles and is in progress. Other equipment to measure different sub-glycans also needs to be developed in order to perform total glycomic analysis. We hope to introduce our analytical studies of another sub-glycan in the future.