
Tadasu Urashima
After graduation from Tohoku University (Doctor of Agriculture) in 1986, he started his professional career by studying milk oligosaccharides at Obihiro University. In 1991, he studied glycosyltransferase activity in lactating mammary glands of the tammar wallaby (marsupial) under Dr. Michael Messer in the Department of Biochemistry, the University of Sydney. Then he completed a comprehensive study of milk oligosaccharides of monotremes, marsupials, and several species of eutherians with Dr. Messer. He is interested in how the present milk components have been acquired during the evolution of mammals, especially how the acquisition of α-lactalbumin, a milk protein, has resulted in the appearance of milk oligosaccharides and lactose, and caused their biological significance to change during evolution. In 2003 ~ 2022, he had been a full professor at Obihiro University, and is an honorary professor today. At present he is a secretary of Japanese Consortium for Glycoscience and Glycotechnology (JCGG).

Kenji Fukuda
Kenji Fukuda received a PhD degree from Hokkaido University in 2002 under the supervision of Professor Atsuo Kimura whose research expertise was in the field of carbohydrate-related enzymes. He worked as a Postdoc in Professor Birte Svensson’s laboratory at Carlsberg laboratory and Technical University of Denmark until the end of 2004. He started his career at Obihiro University of Agriculture and Veterinary Medicine in Japan from 2005, and then appointed to the current position in 2021. His research interests are in exploration, characterization, and application of bioactive substances such as oligosaccharides and peptides found in raw and fermented dairy products, and polysaccharides produced by lactic acid bacteria, under the concept of “One Health”.

Motomitsu Kitaoka
Faculty of Agriculture, Niigata University. Ph.D., Agriculture.
Graduated from Faculty of Engineering, The University of Tokyo in 1985, and after working for a private company, he earned his Ph. D. degree in 1993. From 1995 to 1998, he studied the reaction mechanism of dextran synthase as a postdoctoral fellow at Iowa State University. From 1998 to 2019, he was engaged in research focusing on the development of practical production technology for oligosaccharides using enzymes at the Food Research Institute, National Agriculture and Food Research Organization. He is now a professor at Niigata University since 2019.

Shinya Fushinobu
Professor, Department of Biotechnology, The University of Tokyo
1994 B. S., Faculty of Agriculture, the University of Tokyo; 1996 M. S. Graduate School of Agricultural and Life Sciences, The University of Tokyo; 1997 Assistant Professor at Graduate School of Agricultural and Life Sciences, The University of Tokyo; 1999 Ph.D. Graduate School of Agricultural and Life Sciences, The University of Tokyo; 2006 Visiting Scientist, Iowa State University, U.S.A.; 2011 Promoted Associate Professor at Graduate School of Agricultural and Life Sciences, The University of Tokyo; 2012 Promoted Professor at Graduate School of Agricultural and Life Sciences, The University of Tokyo.
Research interests. Structure and function of enzymes. Structural and functional diversity of Carbohydrate-Active enZymes (CAZymes).

Takane Katayama
Professor, Graduate School of Biostudies, Kyoto University
Takane Katayama is a Professor at Kyoto University. He received his Ph.D in the field of applied molecular microbiology under the supervision of Prof. Hidehiko Kumagai. After spending three years as a postdoc in the same lab, he moved to the lab led by Prof. Kenji Yamamoto and was appointed an assistant professor. In Prof. Yamamoto’s lab, he isolated the genes for 1,2-α-L-fucosidase and endo-α-N-acetylgalactosamindase from bifidobacteria, both of which are enzymes acting on human-derived glycans. These findings prompted him to consider host glycans-mediated symbiosis between gut microbes and humans. In the last decade, he has focused on functional analysis of bifidobacterial genes and enzymes involved in degradation of human milk oligosaccharides. His research has significantly contributed to revealing how bifidobacteria-rich microbiota is formed in the gut of breast-fed infants.
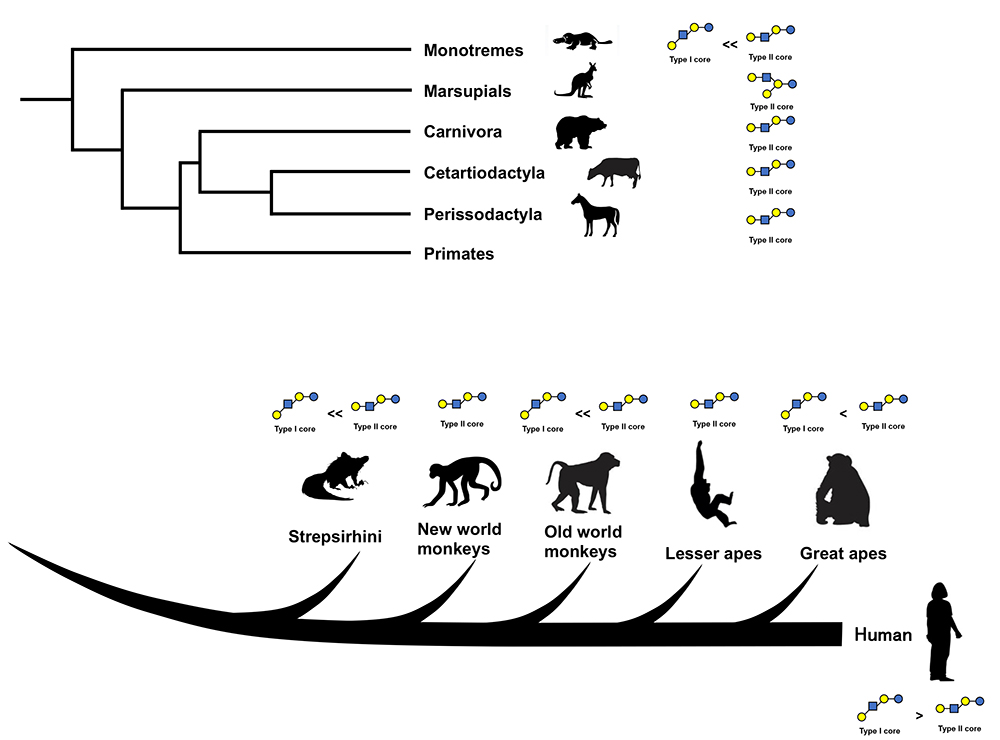
Urashima gave a lecture entitled “The predominance of type 1 oligosaccharides is a feature specific to human breast milk” in May of 2011 at the first human milk oligosaccharides (HMOs) glycobiology meeting held by Glycom A/S (now Royal DSM) in Copenhagen. In this lecture, he proposed that type 1 oligosaccharides are predominant in human milk based on the comparison of the chemical structures as well as profiles of oligosaccharides in human milk with those in the milks of other mammals including non-human primates, carnivora, cetartiodactyla, proboscidea, marsupials, monotremes, etc1. However, as some papers previously maintained that lacto-N-neotetraose (LNnT), a type 2 HMO, is the most dominant oligosaccharide in human milk2,3, his hypothesis was not well accepted. However, the hypothesis of LNnT predominance was based on the peak’s height in the HMO elution profiles on high – performance liquid chromatography (HPLC), even though the LNnT peak was not well separated from that of lacto-N-tetraose (LNT), a type 1 HMO. The quantitative data provided by Thurl et al. (2010), who determined the concentrations of some representative HMOs out of the around 200 known HMOs, showed that the ratios of LNT vs LNnT, as well as the ratios of the saccharides containing LNT core vs those containing LNnT core, were 4-5 fold higher in the former (LNT and saccharides containing LNT core) than in the latter (LNnT and saccharides containing LNnT core) in the breast milk of secretor donors, and higher still in the milk of the non-secretor donors4. As the number of citations of this paper now exceeds 450, the premise should be generally accepted. Thus, the predominance of type 1 HMOs over type 2 is the consensus view at present.
What is the ratio of type 1 milk oligosaccharides (milk OSs) to type 2 milk OSs in non-human mammals? In the milks of the monotremes (platypus and echidna), only one type 1 oligosaccharide containing an LNT core has been found in platypus milk, while some type 2 oligosaccharides containing LNnT or lacto-N-neohexaose (LNnH) core units have been identified in this milk5,6,7. This means that platypus milk is quantitatively and qualitatively richer in type 2 OSs than in type 1 OSs. In the milks of marsupials including tammar wallabies, red kangaroos, common brushtail possums, common wombats, koalas, eastern quolls, and tiger quolls, it has been shown that the oligosaccharides, except for those with a lactose core, have only a core of lacto-N-novopentaose I (novo LNP-I, type 2 OS)5,6,7. Although in the milk carbohydrates of many mammals, lactose predominates in amounts, the milk OSs are more dominant than lactose in the milks of monotremes, marsupials and caniformia other than house dogs5,6,7,8. Even though in the milk of humans, Asian elephants, and African elephants, lactose predominates in amount, milk OSs constitutes 20 ~ 40 % of milk carbohydrates6,9,10. Previously milk OS structures had been characterized using instrumental methods such as nuclear magnetic resonance spectroscopy (NMR) after the separation and purification of each OS, and it is rather easy to characterize them if milk is rich in OSs as is the case in the above mammals. In studies of the milk OSs of bears (including American black bears and polar bears), raccoons, minks, skunks, seals (including hooded seals), Asian and African elephants, Minke whales, etc. Urashima found OSs containing type 2 cores including LNnT, LNnH, para LNnH, but not OSs containing type 1 cores5,6,11,12. However, in these studies, only oligosaccharides with rather high concentrations have been characterized, as NMR-based characterization needs much larger sample volume than mass spectroscopy (MS)-based characterization. The saccharides that exist in small concentrations in the milks may remain uncharacterized. Less abundant type 1 OSs may exist in these milks. Type 1 OSs have been found in the milks of house dogs, house cats. and African lions in other studies by the high-throughput glycome method using MS13,14.
Marino et al. (2010) characterized bovine milk OSs, while Albrecht et al. (2014) characterized the milk OSs of cows, goats, sheep, camels, horses and pigs using HPLC-MS methods after fluorescent labeling in combination with exoglycosidase digestion15,16. It is possible to comprehensively characterize milk OSs including less abundant OSs with their method. Only type 2 OSs with core units LNnT, LNnH, novo LNP-I, or iso LNnT but not type 1 cores, were detected except for the OSs with lactose cores. Remoroza et al. (2020) also did not find type 1 OSs in the milks of cows and buffalos in their study14. A few type 1 OSs have been found in the milks of some domestic farm animals. For example, LNT and LNH were found in goat’s milk14; LNT, LNH, sialyl lacto-N-tetraose a (LST-a), and disialyl lacto-N-tetraose (DSLNT) were detected in mare’s milk17,18, and LNT, lacto-N-fucopentaose II (LNFP-II), LNH, lacto-N-difucohexaose (LNDFH), LST-a, LST-b, sialyl(S)-LNFP-II, S-LNH, and fucosylsialyl(FS)-LNH were identified in sow’s milk19 by capillary electrophoretic separation or HPLC after fluorescent labelling in combination with the MS technique. Interestingly the data obtained by Albrecht et al.16 who did not find type 1 OSs, differed from obtained by Difilippo et al.18,19, who detected type 1 OSs in mare’s and sow’s milks. Although this might have been caused by the difference between the breeds and also between the individual animals in both studies, it is also possible to attribute to the difference in analytical methods used by these two groups. As it is not easy to identify Galβ1-3GlcNAc or Galβ1-4GlcNAc units, and also Galβ1-3(Fucα1-4)GlcNAc or Galβ1-4(Fucα1-3)GlcNAc units by the fragmentation patterns of MS, skilled observation is needed for the identification for these units. During the analytical step, it may be necessary to use digestion by β-galactosidases with different specificities for Galβ1-3GlcNAc or Galβ1-4GlcNAc.
It is certain that only type 2 OSs exist in the milks of these domestic animals other than lactose core OSs, or that type 2 OSs predominate over type 1.
The milks of primate species that are closely related to humans contain both type 1 and 2 OSs. Chimpamzee milk contained LNT (type 1 OS) and LNnT (type 2 OS) in the ratio of 1:1, and LNFP-III (type 2 OS), but not LNFP-I and -II (type 1 OS)20. Bonobo milk contained LNT, LNFP-I as well as LNFP-III, and the ratio of LNFP-I vs LNFP-III was 1:4, showing that the type 2 OSs predominate over the type 120. Gorilla milk contained LNnT but not LNT20. Orangutan milk contained LNT, LNnT, LNFP-III, difucosy LNnH (type 2 OS), LST-b (type 1 OS), LST-c (type 2 OS) and FSLNnH (type 2 OS), and the results of the gel filtration and HPLC analysis of the carbohydrate fraction showed that the type 2 OSs predominate over type 120. Siamang milk contained the type 2 OSs of LNnT, LNnH, LST-c, and SLNnH, but not the type 1 OSs20. Thus, it was shown that the type 2 OSs predominate over the type 1 OSs in the milks of lesser and great apes.
The milks of old world monkeys including rhesus macaques, toque macaques, and Hamadryas baboons, and new world monkeys including tufted capuchins, Bolivian squirrel monkeys and mantled howlers contained type 2 OSs such as LNnT, LNFP-III, novo LNP-I, LNnH, DFLNnH, LST-c and SLNnH, etc, oligosaccharides, but not type 1 OSs21. Among the milks of strepsirrhine species including greater galagos, aye-ayes, Coruerel’s sifakas, and mongoose lemurs, only aye-aye milk contained type 1 OSs such as LNT and LNFP-II, while type 2 OSs predominated over type 1 22. Only type 2 OSs were detected in the milks of other strepsirrhine species22.
These results showed that the milks of some primates contain both type 1 and type 2 OSs, unlike those of some domestic farm animals such as cows, sheep, and camels, though type 2 OSs predominate in the milks of non-human primates. This suggests that the predominance of type 1 OSs in milk is a human-specific feature. Note that because the structural characterization of these OSs was performed by 1H-NMR-based method after the separation of each OS component, the minor oligosaccharides in milk might have not been completely characterized. Since HPLC-MS can detect OSs even at very low levels, it could be used to identify type 1 OSs in the milks of old and new world monkeys and many strepsirrhine species. In another study using the glycome method with HPLC-MS, type 1 OSs as well as type 2 OSs have been identified in the milks of chimpanzees, gorillas, siamangs, rhesus macaques, common marmosets, and golden lion tamarins23. As, up to now, sample sizes have been small, further studies of primate milk OSs should be done with much larger samples.
As above, the notion of some correlation was shown between milk oligosaccharide profiles and the phylogeny of mammalian species. (Fig. 1) However, the completely different data disputing this notion have been obtained for Asian elephant milk by Kunz et al (1999)24, and the milks of twelve species including the gorillas, orangutans, chimpanzees, bonobos, grizzly bears, dogs, Goodfellow tree kangaroos, giraffes, black rhinos, Florida manatees, and bottlenose dolphins by Warren et al. (2001)25, as many type 1 OSs were identified in these milks. These were initial studies characterizing milk oligosaccharides by comparing peak retention times on the HPLC chromatograms of the carbohydrate fractions separated from mammalian milks with those on the HPLC chromatograms of the carbohydrate fractions separated from human milk. As there was less information on non-human milk OS structures, identifications might have been influenced by the bias that the milk OS structures might not be so different between human and other mammals. When we used the NMR technique after each OS separation to characterize the milk OS structures of some mammals other than humans, we noted that some OSs contained unique structural units such as the β1-3 galactosyl linear unit, non-reducing A antigen (GalNAcα1-3[Fucα1-2]Gal), B antigen (Galα1-3[Fucα1-2]Gal) or α-Gal epitope (Galα1-3Galβ1-4GlcNAc), and a core unit containing novo LNP-I. Human milk contained none or only trace amount of these structural units. We believe that structural characterization after OS separation was meaningful in the initial stage of the study of OSs in non–human mammalian milks, even though most recent studies have been using the comprehensive glycome method with HPLC-MS, etc. However, it was difficult for us to collect a large amount of milk from many species other than domestic animals for our studies.
Recently, D. Bojar’s group, using LC-MS/MS analysis in combination with exoglycosidases digestion, characterized around 400 OSs including 100 novel structures in the milks of nine mammalian species including alpacas, beluga whales, black rhinoceroses, bottlenose dolphins, impalas, L’Hoest’s monkeys, pygmy hippopotamuses, domestic sheep, and striped dolphins26. Their data significantly expanded the information on the diversity of milk OS structures. Noteworthily, OSs containing the GalNAcβ1-4GlcNAc (LacdiNAc) motif were found in all the milks except those from domestic sheep and striped dolphins, and the OSs containing glucuronic acid (GlcA) were detected in milks from impalas, bottlenose dolphins, striped dolphins, domestic sheep, and black rhinoceroses. Free LacdiNAc was previously identified in bovine milk, which was the only milk OS with this OS motif15, found in the previous studies. The milk OS containing the core units of LdiNnT and LdiNnH, which are related structures of LNnT and LNnH, were identified in the milks of impalas and pygmy hippopotamuses. Also noteworthy is the finding of OSs containing LacdiNAc in the milk of L’Hoest’s monkeys (a old world monkey) as well as in the milks of cetartiodactyla species. During the biosynthesis of lactose, the acceptor specificity of β4GalT1 is changed from GlcNAc to Glc as a result of its association with α-lactalbumin, a milk protein27, while the donor specificity of β4GalT1 has also been shown to change from UDP-Gal to UDP-GalNAc in in vitro experiments27,28. The LacdiNAc motif of milk OSs, found in the milks of these species might have been biosynthesized by molecular substrate wobbling, caused by such association of β4GalT1 with α-lactalbumin.
The OSs containing Sda epitope (Neu5Acα2-3[GalNAcβ1-4]Gal) were identified in the milks of beluga whales, bottlenose dolphins, and black rhinoceroses26. Although the smallest OS containing this epitope, Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glc (designated the GM2 tetrasaccharide) had previously been found in the milks of bottlenose dolphins29, rhesus macaques21 and giraffes30, it was first that the larger OSs, containing Sda epitope, with LNnT, LNnH, LdiNnT, or LdiNnH core units were identified in addition to the GM2 tetrasaccharide. Only the smallest one could have been identified by Urashima’s group, who used NMR for characterization after each OS separation. A breakthrough was achieved by the development of the glycome analysis method, which can be used to determine OS structures in a very small amount of milk. The technique of OS characterization has been dramatically improved by the accumulation of empirical information on fragmentation patterns specific to some structural units in the MSn spectrum, in combination with digestion using substrate-specific exoglycosidases. It is speculated that precise milk OS profiles will be obtained in many mammalian species in the near future; this will clarify the large heterogeneity of milk OS structures in mammals. The development of methods of characterization of OS structures in non–human mammalian milks is historically similar to that of HMOs. Until the 1970s, the classical methods of monosaccharide analysis, methylation analysis, and exoglycosidase digestion were used to determine HMOs structures; then from 1980s to 2000, 1D and 2D NMR-based techniques were utilized for the characterization. The last identification with NMR was done for lacto-N-decaose (LND) and fucosyl lacto-N-decaose I (F-LND-I) by Chai et al (2000)31, and MSn-based methods were thereafter for HMO structure characterization32,33,34,35,36.
Even though the structural diversity of milk OSs among mammals was dramatically expanded by Jin et al.26, present findings of the distribution of the ratios of type 1 to type 2 milk OSs among mammals should not differ from previous observation (Fig. 1) except for the milk of L’Hoest’s monkey. L’Hoest’s monkey milk rarely contains type 1 OSs of LST-b (type 1 OS) as the second most dominant saccharide after lactose. As Goto et al. identified LST-c (type 2 OS), but not LST-b, in the milks of rhesus macaques and Hamadryas baboons13, which are closely related species to L’Hoest’s monkey, the L’Hoest’s monkey may be unique among the old world monkeys for producing milk with an abundance of LST-b.
A comprehensive glycan database for milk OSs (MilkOligoDB) has been updated and is now available37, and the correlation and evolution of milk OS structures among mammalian lineages as well as the potential of some biosynthetic pathways have been hypothesized37,38. Thomes et al. (2023) demonstrated that marsupial milk OSs as uniquely grouped and diverge from the other milk OSs38, though Warren et al.’s data of milk OSs of the Goodfellow tree kangaroos25 largely differ from other marsupial milk OSs, which have structures that are interrelated among the species. As described above, when discussing the evolution of milk OSs in relation to mammalian phylogeny, we suggest that Warren’s milk OS data of twelve species be excluded from the milk OS database.
Since the first HMOs glycobiology symposium in 2011, information about milk OS structures in mammals other than humans has dramatically expanded. Information about the milk OSs of cows, goats, sheep, camels, horses, and pigs has been extended by Albrecht et al. (2014)16, that of cows, buffalos, goats, and lions has been developed by Remoroza et al. (2020)14, and that of nine mammalian species has been developed by Jin et al.26. It is likely that the hypothesis of type 1 milk OS predominance specific to humans should have been supported by these studies.
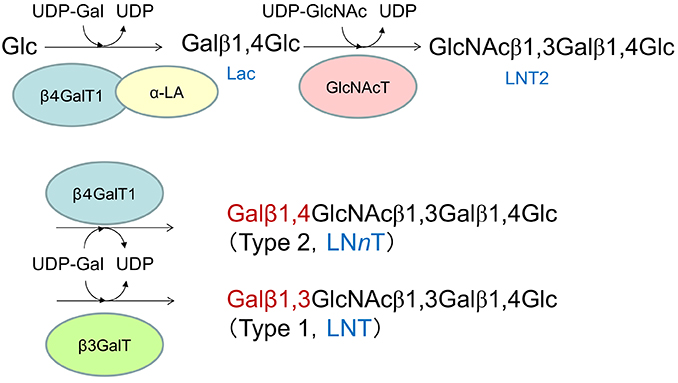
Type 1 and type 2 OSs are biosynthesized from their respective core tetrasaccharides (LNT and LNnT) as the starting compounds. These core tetrasaccharides are generated through galactosyl-transfer by different galactosyltransferases to lacto-N-triose II (LNT2), which is formed by the transfer of GlcNAc to the non-reducing terminal Gal residue of lactose at OH-3 (Fig. 2). The biosyntheses of LNT and LNnT by the two β-galactosyltransferases are, thus, competitive reactions sharing LNT2 and UDP-Gal as the substrates. Because β4GalT1 is the same enzyme involved in lactose biosynthesis, it is considered to be expressed in large quantities in the mammary glands of species expressing milk with lactose as the main carbohydrate. Therefore, the higher activity of β3GalT than β4GalT1 is required to actualize the type 1 OS predominance in milk (Fig. 2).
Compared to cows, humans have around 100 times higher concentration of milk OSs and much wider variety of milk OSs. It is hypothesized that the evolution of HMOs has been directed to greater variety, higher concentration, and the predominance of the type 1 over type 2. Recently, the number of publications on the biological functions of HMOs has significantly increased, and the relationship of the biological functions of HMOs to stimulation of the growth and colonization of beneficial colonic bifidobacteria in the breastfed infant colon, anti–infection activity against the pathogenic viruses and bacteria, immune modulation, prevention for necrotizing enterocolitis, the activation of infant brain activity, etc,30 has been noted. It is assumed that some performance of these functions depends on the actions of the products of HMO metabolism by colonic bifidobacteria. In the last 1.5 decades, the metabolic pathways of type 1 HMOs have been clarified mainly by groups in Japan and at the University of California Davis. As the result of acquisition of the predominance of type 1 milk OSs by humans as well as the metabolic pathway of the type 1 HMOs by human infant-type bifidobacteria, the symbiotic evolution between humans and colonic bifidobacteria has been occurring; this should play a significant role in the healthy development of the human breastfed infants. We will concisely explain how infant-type bifidobacteria have acquired the ability to metabolize type 1 HMOs.
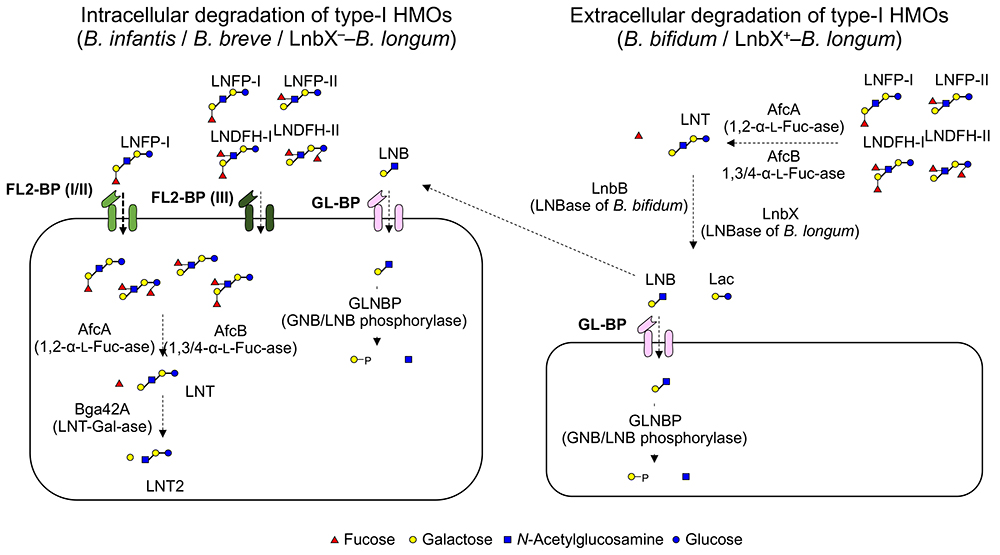
The predominant growth of bifidobacteria in the intestinal tract of breastfed infants has been known since the beginning of the 20th century. Around 1960, it was assumed that the HMOs in breast milk acted as growth factors for bifidobacteria. The discovery of the β-1,3-galactosyl-N-acetylhexosamine phosphorylase (GLNBP) in the cells of Bifidobacterium bifdum40 that specifically phosphorolyzes lacto-N-biose I (LNB; a structural disaccharide unit in the type 1 OSs) has rapidly advanced our understanding of the HMO metabolic system of bifidobacteria. Since the GLNBP gene was identified from multiple species of bifidobacteria, the LNB hypothesis was proposed, explaining the preferable growth of bifidobacteria by the selective utilization of LNB released from the non-reducing end of type 1 OSs41 (Fig. 3 right). The GLNBP gene is widely distributed in major infant-resident bifidobacterial species, but little of it is distributed in others42. In addition, the bifidobacterial species fermenting LNB were almost limited within that possessing the GLNBP gene. Based on the LNB hypothesis, the extracellular enzymes required to release LNB from type I OSs (including fucosidases, sialidases, and lacto-N-biosidase) were identified in B. bifidum43. However, advanced genomic analysis has revealed that B. bifidum is the only bifidobacterial species possessing such an extracellular enzyme system44.
Although B. bifidum is the sole species that possesses the gene set required for digesting HMOs extracellularly into mono- and disaccharides, an extracellular lacto-N-biosidase, that belongs to GH136, has been identified from Bifidobacterium longum subsp. longum45. GH136 lacto-N-biosidase is present in one-third to half of B. longum strains, and acts not only on LNT but also on LNFP-I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and LST-a (Neu5Acα2-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc). The presence of GH136 lacto-N-biosidase and its cognate chaperon in several B. longum strains might also represent the adaptation of bifidobacteria to type 1 HMOs. It is interesting to note that B. bifidum and B. longum have evolved two lacto-N-biosidases distinct in amino acid sequences despite that they colonize the same niche.
Bifidobacterium longum subsp. infantis has adopted a different strategy for digesting type 1 HMOs. A specific characteristic of B. infantis is the presence of a large gene cluster (43 kbp, termed HMO cluster I) consisting of many glycosidases and transporters involved in the HMO metabolism46. However, the β-galactosidase encoded in this cluster is specific for type 2 HMOs and does not hydrolyze type 1 HMOs. Yoshida et al. found that a GH42 β-galactosidase (termed Bga42A), that is encoded at a different locus from HMO cluster I, is responsible for type 1 HMOs47. B. infantis has three GH42 β-galactosidases including Bga42A. It is noteworthy that Bga42A is also able to hydrolyze Galβ1-3Galβ1-3Galβ1-4Glc48, which suggests that β-galactosidases with affinity for Galβ1-3Gal linkage has acquired an ability to hydrolyze type 1 structure present in LNT (Fig.3 left).
Bifidobacterium animalis subsp. lactis Bl-04 possesses an β-galactosidase, termed Gal42A, which is a homolog of Bga42A with 62% identity. The enzyme indeed acts on Galβ1-3Gal disaccharide; however, it neither acts on LNB, LNT, Galβ1-3GalNAc (GNB), Galβ1-3Galβ1-4Glc, nor Galβ1-3Galβ1-3Galβ1-4Glc49. These findings suggest that GH42 β-galactosidases need to acquire two abilities to hydrolyze LNT, i.e. one is accommodation of bulky N-acetyl group of GlcNAc or GalNAc near active site and the other is accommodation of sugar chains of ≥ 3 in the catalytic pocket.
Overall, from an enzymatic viewpoint, B. bifidum and B. infantis could represent evolutionary adaptation to type 1 HMOs. These traits and the LNB utilization ability commonly found in infant gut-associated bifidobacteria including B. breve and B. longum contribute to the bifidobacteria-rich microbiota formation in the breastfed infant gut50,51.
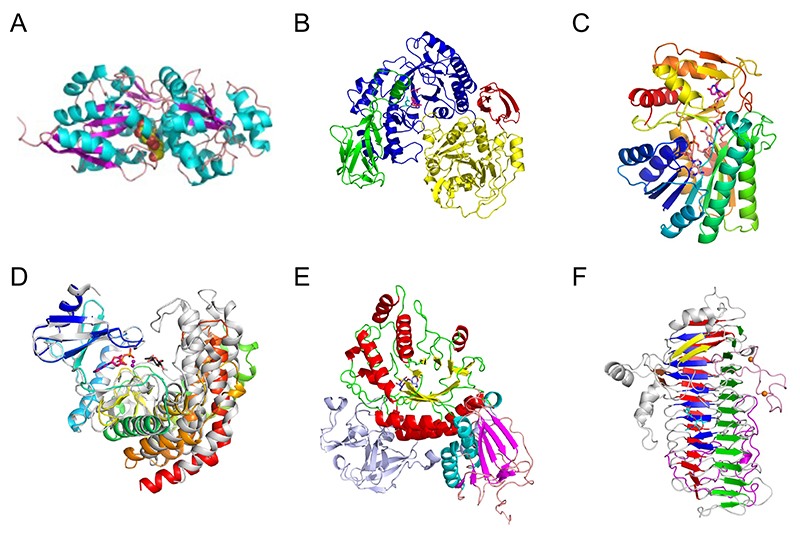
Three-dimensional structures of proteins are often conserved among different families, providing a clue for what kind of molecular evolution they have undergone. Among the enzymes and proteins involved in the metabolic pathway of bifidobacteria specific for type 1 OS, the first three-dimensional structure was reported for the substrate-binding protein of the ABC transporter that imports LNB into the bacterium (Fig. 4A)52. The substrate-binding protein is from B. longum, and it is called GNB/LNB-binding protein (GL-BP) because it also strongly binds GNB. GL-BP extensively recognizes the entire LNB disaccharide, suggesting that this protein has undergone molecular evolution to adapt to the uptake of type 1 OSs. The overall structures of substrate-binding proteins for carbohydrates, including the well-known maltodextrin binding protein (MBP), are all similar and consist of two large domains at the N- and C-lobes. Therefore, through mutations around the substrate-binding site, they likely have evolved to permit uptake of diverse carbohydrates. GL-BP from B. longum binds weakly to the tetrasaccharide LNT, whereas its homolog from B. infantis binds LNT as strongly as LNB. The specificity of the transporter seems to be fine-tuned for each Bifidobacterium species according to the repertory of extracellular enzymes.
Subsequently, the three-dimensional structure of GLNBP, which phosphorylates LNB in the cell, was also reported (Fig. 4B)53. GLNBP belongs to GH112, and its (β/α)8 barrel catalytic domain undergoes a large conformational change upon substrate binding. The overall structure of GLNBP resembles GH14 β-amylase. Therefore, a molecular evolutionary relationship can be inferred between GH112 and GH14, while the carbohydrate structures of α-1,4-glucan (starch) and type 1 OS are quite different. Moreover, the reaction mechanism of these enzymes differs between hydrolysis and phosphorolysis, suggesting that substrate recognition and catalysis (phosphate-binding) of GLNBP may have evolved in a manner involving movement of the catalytic domain. The enzymes in the downstream genes further metabolize the reaction products of GLNBP, galactose 1-phosphate (Gal-1P) and GlcNAc. These metabolic enzymes are similar to those in the Leloir pathway of galactose metabolism. Among them, the epimerase (GalE), which interconverts UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc, has similar sequence and structure to those of the GalE enzymes from Escherichia coli and humans (Fig. 4C)54. However, N-acetylhexosamine kinase (NahK), which phosphorylates the anomeric position of GlcNAc using ATP, and sugar 1-phosphate uridylyltransferase (GalT), which transfers the sugar between sugar 1-phosphates (such as Gal-1P and GlcNAc-1P) and UDP-sugars, have distinct amino acid sequences from known Leloir pathway enzymes. In particular, the three-dimensional structure of NahK from B. longum resembles protein kinases and aminoglycoside glycosyltransferases (Fig. 4D), suggesting a molecular evolutionary relationship among them55.
Lacto-N-biosidase extracellularly cleaves LNB from type 1 OSs. In B. bifidum, a GH20 enzyme (LnbB) is used (Fig. 4E)56. The most abundant GH20 members are exo-type β-N-acetylhexosaminidases that releases monosaccharide (GlcNAc or GalNAc) from the non-reducing end. Thus, LnbB likely evolved from GH20 β-N-acetylhexosaminidase. LnbB may have evolved to have a pocket that accepts β1,3-linked Gal so that it can release the disaccharide. On the other hand, as mentioned earlier, the lacto-N-biosidase in B. longum is a GH136 enzyme (LnbX), which has a unique β-helix structure in contrast to the many enzymes and proteins shown so far to degrade and metabolize type 1 OSs (Fig. 4F)57. While the three-dimensional structure of LnbX has little homology to known enzymes, it is similar to phage tailspike proteins. Some of these tailspike proteins have carbohydrate-binding ability. It was suggested that phage-derived sequences in the bifidobacterial genome (prophage) may have influenced molecular evolution and diversity in the symbiosis with humans58.
After the molecular evolution of α-lactalbumin from lysozyme, the biosynthesis of milk oligosaccharides must have begun along with the beginning of lactose biosynthesis5. It is hypothesized that milk oligosaccharides should have been the dominant saccharide in the primitive milk-like secretions of the common ancestor of living mammals, inasmuch as monotremes, which have the ancestral morphological features of mammals, produce milk containing predominantly oligosaccharides over lactose5,6,7. Among the mammalian species in which milk oligosaccharides have been studied, most species, other than the primates, express milks containing many varieties of type 2 OSs and only trace levels of or no type 1 OSs1. Even though the milks of primates have both of type 1 and 2 milk OSs, type 2 predominates over type 1 in the milks of non-human primates1,23. In human milk, type 1 OSs predominate over type 2, which is thought to be a human-specific feature; in addition, that the concentration of milk OSs is much higher in human milk than in the milks of cows, or goats1,4.
Because colonic bifidobacteria metabolize HMOs and promote the health of human infants, symbiosis between humans and these bacterial species has started. It is thought that the acquisition of the ability to metabolize predominant type 1 HMOs is the key to potential colonization by human infant-type bifidobacteria. It is concluded that this ability was due to the acquisition of affinities for type 1 HMOs and LNB in the transporters, glycosidases, and phosphorylases involved in the HMOs metabolism.
The biological functions of HMOs include protection against pathogenic microorganisms in the colon, immune modulation, prevention of necrotizing enterocolitis, stimulation of brain activity, and enhancement of the colonic barrier39. It is assumed that some functions may be due to the activities of the metabolic products of HMOs in bifidobacteria. There is no doubt that colonic bifidobacteria have definite effects on the survival of human newborns, who are less mature at birth than the newborns of other primates including great apes. As it has been recently shown that the metabolic products produced by colonic bacteria may affect brain activity via the vagus nerve of the gut-brain axis, it is possible that the colonic bifidobacteria have an important effect on the development of the brain in human neonates. If so, it is speculated that the acquisition of type 1 HMO predominance in milk has been a driver of the symbiotic relationship of human with colonic bifidobacteria, which would further advance for the evolution of humans themselves. Today a few HMOs including 2’FL and LNnT, which are manufactured on an industrial scale, are included in milk replacers produced from cow’s milk. We emphasize that the type 1 HMOs such as LNT, LNFP-I, LNDFH-I, and DSLNT will be manufactured and incorporated as ingredients in the infant formula.
lacto-N-triose II (LNT2) : GlcNAcβ1-3Galβ1-4Glc
lacto-N-tetraose (LNT) : Galβ1-3GlcNAcβ1-3Galβ1-4Glc
lacto-N-neotetraose (LNnT) : Galβ1-4GlcNAc1-3Galβ1-4Glc
lacto-N-hexaose (LNH) : Galβ1-3GlcNAcβ1-3(Galβ1-4GlcNAcβ1-6)Galβ1-4Glc
lacto-N-neohexaose (LNnH) : Galβ1-4GlcNAcβ1-3(Galβ1-4GlcNAcβ1-6)Galβ1-4Glc
lacto-N-novopentaose I (novo LNP-I) : Galβ1-3(Galβ1-4GlcNAcβ1-6)Galβ1-4Glc
para lacto-N-neohexaose (para LNnH) : Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc
iso lacto-N-neotetraose (iso LNnT) : Galβ1-4GlcNAcβ1-6Galβ1-4Glc
sialyl lacto-N-tetraose a (LST-a) : Neu5Acα2-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc
sialyl lacto-N-tetraose b (LST-b) : Galβ1-3(Neu5Acα2-6)GlcNAcβ1-3Galβ1-4Glc
sialyl lacto-N-tetraose c (LST-c) : Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc
disialyl lacto-N-tetraose (DSLNT) : Neu5Acα2-3Galβ1-3(Neu5Acα2-6)GlcNAcβ1-3Galβ1-4Glc
lacto-N-fucopentaose I (LNFP-I) : Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc
lacto-N-fucopentaose II (LNFP-II) : Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4Glc
lacto-N-fucopentaose III (LNFP-III) : Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4Glc
lacto-N-difucohexaose I (LNDFH-I) : Fucα1-2Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4Glc
lacto-N-difucohexaose II (LNDFH-II) : Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4(Fucα1-3)Glc
lacto-N-decaose (LND) : Galβ1-3GlcNAcβ1-3{Galβ1-3GlcNAcβ1-3[Galβ1-4GlcNAcβ1-6]Galβ1-4GlcNAcβ1-6}Galβ1-4Glc
fucosyl lacto-N-decaose I (F-LND-I) : Galβ1-3GlcNAcβ1-3{Galβ1-3[Fucα1-4]GlcNAcβ1-3[Galβ1-4GlcNAcβ1-6]Galβ1-4GlcNAcβ1-6}Galβ1-4Glc
lacto-diN-neotetraose (LdiNnT) : GalNAcβ1-4GlcNAcβ1-3Galβ1-4Glc
lacto-diN-neohexaose (LdiNnH) : GalNAcβ1-4GlcNAcβ1-3(GalNAcβ1-4GlcNAcβ1-6)Galβ1-4Glc
2'-fucosyllactose (2'FL) : Fucα1-2Galβ1-4Glc