
Yoshitaka Sato
Associate Professor, Department of Virology, Nagoya University Graduate School of Medicine
In 2004, I graduated from Nagoya University School of Engineering. In 2013, I graduated from Kobe University School of Medicine (college transfer) and completed a shortened doctoral course in the Nagoya University Graduate School of Medicine. After residency at Nagoya University Hospital, I became an Assistant Professor in 2015, Lecturer in 2020, and have been an Associate Professor since April 2022 at Nagoya University Graduate School of Medicine. Also, I have been a JST PRESTO Researcher since 2019. I am presently studying the diverse fates of virally infected cells, focusing on Epstein-Barr virus infected cells.
Many viral pathogens utilize heparan sulfate as attachment factors, which facilitates the initial interaction with host cells. We recently identified 3'-Phosphoadenosine 5'-Phosphosulfate Synthase 1(PAPSS1)as a host factor for herpes simplex virus type 1 (HSV-1) infection by a genome-wide loss-of function CRISPR screen1. The Knockout (KO) of PAPSS1 reduced heparan sulfate expression. Here, I would like to introduce and discuss the role of heparan sulfate on viral infection.
HSV-1 causes various diseases such as labial herpes, genital herpes, and serious encephalitis, and is a clinically problematic virus regardless of immune status. The prevalence of anti-HSV-1 antibody in adults is thought to be 70% or higher, indicating that HSV-1 is a highly prevalent human pathogen. A characteristic of HSV-1 is its broad cell tropism (enabling infection of many cells). The proliferation of HSV-1 exerts a cytopathic effect leading to cell death. Utilizing this character, we performed CRISPR screening to exhaustively identify host factors involved in HSV-1 infection1.
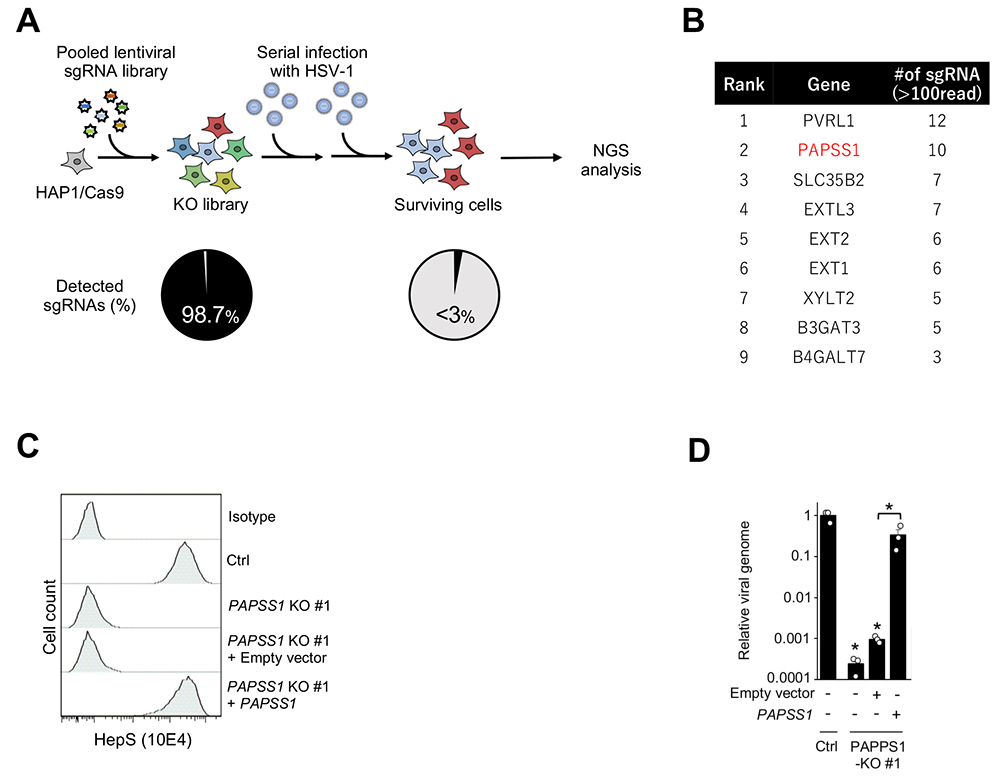
In this screening, we intended to identify HSV-1 host factors by establishing a knockout library encompassing approximately 19,000 genes and 2,000 miRNAs, using HSV-1 to infect cells containing the knocked-out genes and miRNAs in the established library, collecting cells that were resistant to infection, and analyzing sgRNA sequences using next-generation sequencing (Fig. 1A). As HSV-1 host factors, we isolated 11 genes and 1 miRNA including the NECTIN1 gene encoding a virus receptor that is thought to be essential for HSV-1 infection. In addition to nectin cell adhesion molecule 1 (NECTIN1), and xylosyltransferase 2 (XYLT2) and exostosin glycosyltransferase 2 (EXT2), both of which biosynthesize heparan sulfate, we identified 3'-phosphoadenosine 5'-phosphosulfate synthase 1 (PAPSS1), the enzyme that synthesizes PAPS (the only sulfate group donor in mammals), as a novel HSV-1 host factor by validation experiments demonstrating that high resistance to HSV-1 infection was obtained by the KO of the PAPSS1 gene (Fig. 1B).
Since many of the HSV-1 host factors identified by our CRISPR screening are involved in the biosynthesis of heparan sulfate, and sulfation of sugar by sulfotransferase is essential for the biosynthesis of heparan sulfate, we first considered the possibility that PAPSS1 is involved in the biosynthesis of heparan sulfate. Actually, a marked decrease in the expression of heparan sulfate on the cell surface was observed with the KO of PAPSS1 (Fig. 1C), and similar results were obtained by the disaccharide analysis of heparan sulfate sugar chains, indicating the involvement of PAPSS1 in the sulfation of sugar chains and the importance of PAPSS1 in the biosynthesis of heparan sulfate. The attachment of HSV-1 to the cell surface also decreased by a factor of 100–1,000 in PAPSS1-KO cells (Fig. 1D). Therefore, I think that PAPSS1 enables efficient attachment of the virus to cells by contributing to the biosynthesis of heparan sulfate1.
The attachment of HSV-1 to the surface of target cells mediated by heparan sulfate has been reported since the 1980s2, and it is known that viral glycoproteins gB and gC bind to heparan sulfate3. This binding is involved in the binding of viral particles to the host cell surface but not in the entry of viral particles into the host cell. The entry of HSV-1 requires the binding of the glycoprotein gD to entry receptors and subsequent fusion of the viral envelope with the host cell membrane. In addition to NECTIN-1, NECTIN-24, and HVEM (herpesvirus entry mediator)5, 3-O-sulfated heparan sulfate has been identified6 as an entry receptor. Interestingly, only 3-O-sulfated heparan sulfates sulfated by human 3-O-sulfotransferases 3-OST-2, 3-OST-3A, 3-OST-3B, 3-OST-4, 3-OST-5, 3-OST-6, but not 3-OST-1 interact with gD and function as entry receptors6. Although it is not yet clear why 3-O-sulfated heparan sulfate that is sulfated by 3-OST-1 cannot serve as an entry receptor, it is not difficult to imagine that more than one sulfotransferase acting in concert participate in sulfation and produce sugar chains with more complex sulfation patterns. Such functional redundancy of enzymes was also a weakness in the loss-of-function CRISPR screening described above, and we were not able to isolate OSTs as HSV-1 host factors. To isolate such enzymes, a gain-of-function screening needs to be performed. When 3-O-sulfated heparan sulfate was identified as a receptor for gD, an ORF expression library was used. Thus, effective utilization of a gain-of-function screening with CRISPRa may enable more extensive elucidation of the relationship between viral particles and sugar chains.
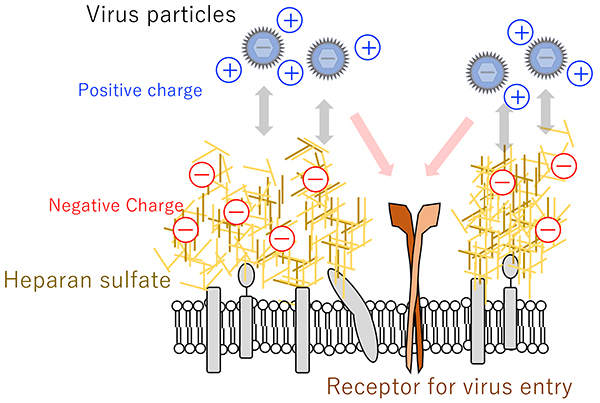
Besides HSV-1, a wide variety of viruses have been reported to interact with heparan sulfate including human immunodeficiency virus7, hepatitis B virus8, dengue virus9, human papillomavirus10, common cold coronavirus11, vaccinia virus12, and Lassa virus13. Therefore, the electrostatic interaction between positive charges on the surface proteins of viral particles and negative charges on heparan sulfate molecules is thought to be involved in viral particle attachment to the cell surface14,15 (Fig. 2).
SARS-CoV-2, the causative virus of coronavirus disease 2019 (COVID-19), which originated in Wuhan City, China, in December 2019 and subsequently developed into a pandemic, also utilizes heparan sulfate for binding to the surface of target cells11,16. The spike protein (S protein) present in the envelope of SARS-CoV-2 binds to the ACE2 receptor present on the cell membrane, and the virus enters the cell. In this process, the S protein also binds to heparan sulfate and this interaction strengthens the binding to the ACE2 receptor16.
Since heparan sulfate is abundant on the cell surface, I think it is fitting that viruses utilize heparan sulfate as a scaffold for attachment to target cells. On the other hand, it is known that heparan sulfate structures are diverse and vary among cell types, from individual to individual, by age, and by other factors17-20. It is possible that this structural diversity of heparan sulfate determines tropism and susceptibility to virus infection, and it is interesting that deeper analysis of the interaction between viral particles and heparan sulfate may lead to better understanding of diverse heparan sulfates.
The screening for HSV-1 host factors presented in this article was collaborative research with Takeshi Suzuki, PhD (graduate student at that time) in the Department of Virology, Nagoya University Graduate School of Medicine (under Professor Hiroshi Kimura), Yusuke Okuno, PhD at Nagoya City University, Hiroshi Kitagawa, PhD at Kobe Pharmaceutical University, and others, and was conducted with grant support from KAKENHI (JP16H06231, JP19H04829, JP21K15448, JP20K06551, JP20H03386, and JP20H03493), JST (JPMJPR19H5), AMED Interdisciplinary Cutting-edge Research (JP21wm035042 and JP20wm0325012), and others.