Hideo Mochizuki
Hideo Mochizuki graduated from the Faculty of Science, Konan University (1983) and obtained his Ph.D. from the Graduate School, Kanazawa University (1990). In 1990, he entered Seikagaku Corporation and retired in 2020. He served as Visiting Researcher in the Institute for Molecular Science of Medicine, Aichi Medical University, directed by the lifelong master Professor Koji Kimata (2002-2004). He is currently a contract employee at Seikagaku Corporation. He first became aware of non-AT-binding 3S in 2000, and has continued to work in this field for 20 years.
Heparan sulfate (HS) is present as a component of proteoglycans on cell surfaces and in the extracellular matrix in most animal species including Hydra, Caenorhabditis, Drosophila, and humans. HS chains are structurally heterogeneous, being composed of densely sulfated regions, or sulfated domains, connected by mostly nonsulfated and N-acetyl-rich regions. HS regulates various physiological processes by the interaction with numerous proteins such as growth factors, morphogens, cytokines, enzymes, and extracellular matrix proteins. The binding specificity of HS for each functional protein is thought to be dependent on the structure of the sulfated domain.
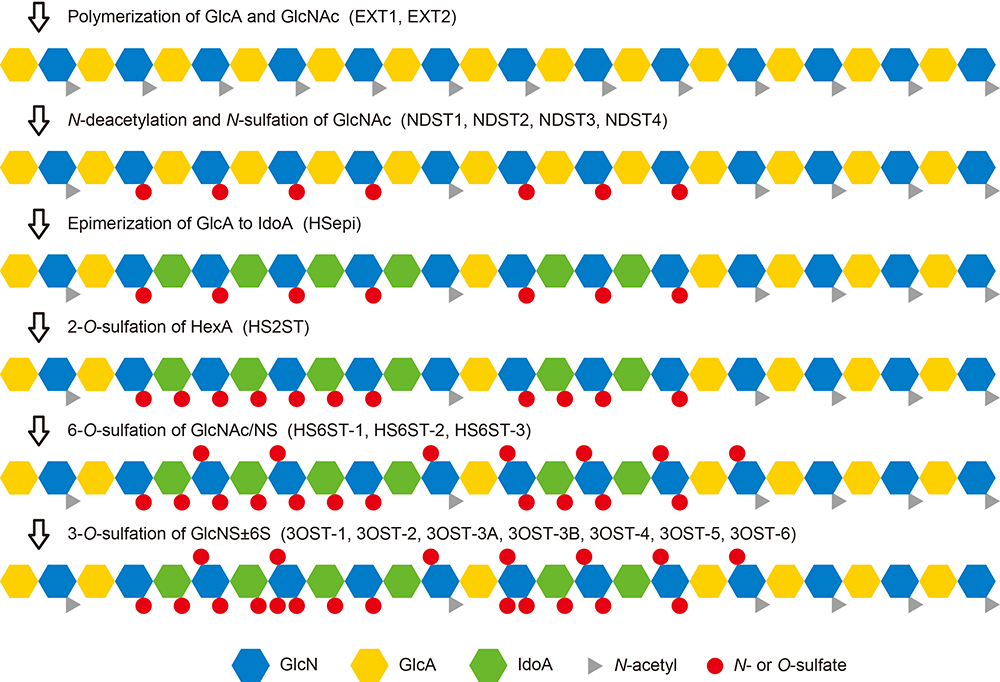
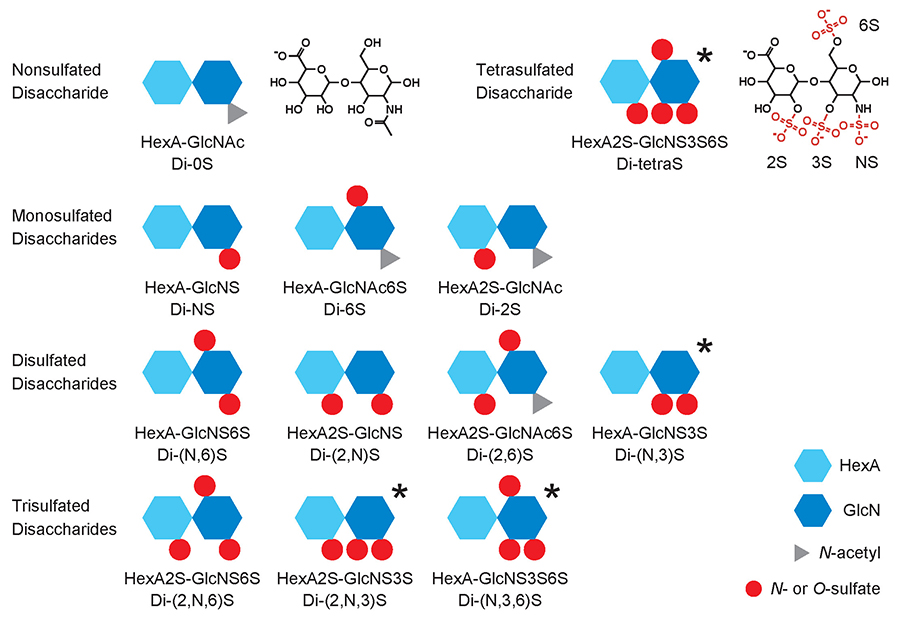
HS is synthesized by a group of enzymes located in the Golgi apparatus, by the pathway shown in Figure 1. The several reactions of the pathway are catalyzed by multiple isoforms as shown inside parentheses. The biosynthesis begins with the formation of a repeating disaccharide structure, -GlcA-β1,4-GlcNAc-α1,4-, by alternate addition of glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) residues. Next, part of the region on the linear chain undergoes N-deacetylation/N-sulfation, forming the base for the sulfated domain. In addition, GlcA residues undergo isomerization to form iduronic acid (IdoA), the hexuronic acid (HexA) residues undergo 2-O-sulfation, and glucosamine residues undergo 6-O-sulfation. Another reaction that sometimes occurs, albeit rarely, is 3-O-sulfation of glucosamine residues. Because these modification steps are heterogeneous, disaccharide units with a variety of modified structures are formed. The 12 sulfation patterns that occur naturally are shown in Figure 2. Some of these disaccharide units have HexA isomers, i.e., GlcA or IdoA, resulting in an even wider variety of structures. Linear arrangement of these makes an enormous number of possible combinations, and it is possible that the HS bind specifically to individual functional proteins. Another way of looking at this is that the specific arrangements of sulfate groups on HS chains form structures that are key for specific physiological activities. Therefore, rare disaccharide or heavily sulfated disaccharide units are particularly important components for achieving key structures.
From the above perspective, an important area of research is elucidation of the functions of the 3-O-sulfated structure (3S). Although disaccharide units with 3S are usually less than 1% of the total disaccharides content, mammals including humans have seven isoform genes for an enzyme that catalyzes this rare modification reaction (Fig. 3). It was considered necessary to obtain many isoforms with different specificities, in order to recognize different surrounding structures, and achieve 3-O-sulfation of glucosamine residues.

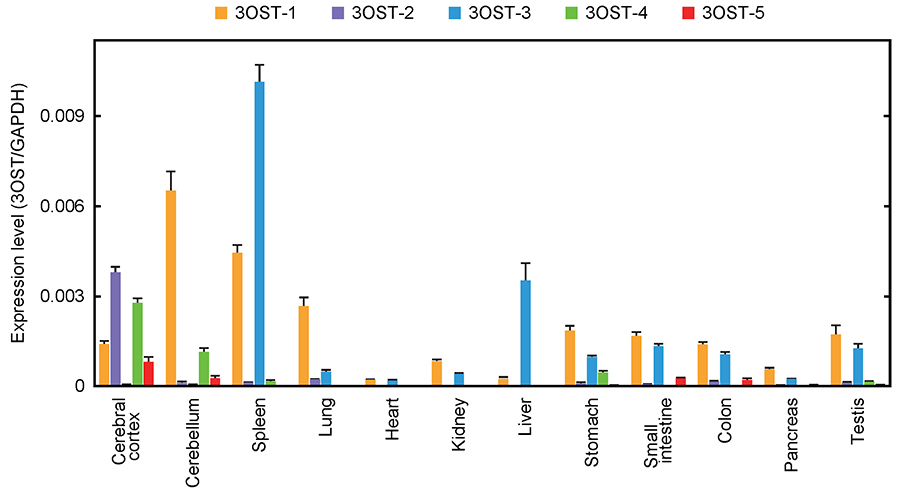
Research on 3S has been in progress for a long time. In 1980, Lindahl et al. first identified 3S as the critical component of heparin binding to antithrombin (AT)1. Because of the importance of heparin for medical treatment, thorough research has been performed on this AT-binding 3S2. The pentasaccharide shown in Figure 4 has the minimum structure needed for binding with AT, and AT-binding ability is lost when the central 3S is eliminated3. The enzyme needed for the synthesis of AT-binding 3S, 3-O-sulfotransferase 1 (3OST-1), has been purified and cloned4,5. Whereas 3OST-1 was identified on the basis of physiological function, the other six isoforms were identified in the DNA database as genes homologous with the 3OST-1 gene6-8. All these genes have been cloned, and the enzymatic properties have been investigated in vitro. Interestingly, unlike 3OST-1, five of the isoforms, 3OST-2, 3OST-3A, 3OST-3B, 3OST-4, and 3OST-6, do not form AT-binding 3S. 3OST-5 forms both AT-binding and non-AT-binding 3S. Little progress has been made with elucidation of the physiological functions of these non-AT-binding 3Ss. Figure 5 shows the expression levels of 3OST isoforms in human tissues. Whereas 3OST-1 and 3OST-3 show relatively widespread expression, 3OST-2 and 3OST-4 are specific to the central nervous system. The expression levels of each isoform vary considerably, and tissue-specific expression patterns can be identified.

Disaccharide analysis is in general use for compositional analysis of HS. Three types of heparin lyase have been isolated from Flavobacterium heparinum, which grows using heparin as a nutrient source. These differ in specificity to sulfation structures, and a mixture of the three lyases enables almost complete cleavage of the glucosaminidic linkages in HS, with one exception. In the digestion products, a double bond has been introduced to HexA between C4 and C5, forming an unsaturated disaccharide and thereby losing isomer information, i.e., GlcA or IdoA. Only one of the structures is resistant to lyase-digestion: the AT-binding structure. The glucosaminidic linkage in the non-reducing terminal of GlcA-GlcNS3S±6S units is resistant to digestion and results in unsaturated tetrasaccharides9, whereas the glucosaminidic linkage adjacent to IdoA±2S-GlcNS3S±6S units (non-AT-binding 3S) is susceptible to the lyases and digested into unsaturated disaccharides10. Separation and quantification of the digestion products by high-performance liquid chromatography (HPLC) has shown the disaccharide composition of HS.
We digested the reaction products of 3OST isoforms using heparin lyases, and then separated and identified the five 3S components. Of these five, three were disaccharides (non-AT-binding 3S), these being Di-(N,3,6)S, Di-(2,N,3)S, and Di-tetraS units. The other two components were lyase-resistant tetrasaccharides, which were provisionally named Tetra-1 and Tetra-2, and GlcA-GlcNS3S and GlcA-GlcNS3S6S, respectively, were found at their reducing terminals. Figure 6 shows the results of an investigation relating to each isoform, for the production ratios of 3S components formed by sulfation of HS and heparin (external and internal donut charts, respectively). In the case of 3OST-1, two tetrasaccharides were the principal products, whereas almost no tetrasaccharides were formed from 3OST-2, 3OST-3, or 3OST-4, and the principal products were Di-(2,N,3)S and Di-tetraS units. Unlike other isoforms, 3OST-5 forms both disaccharides and tetrasaccharides, and in addition shows a marked difference in specificity for the two substrates. Disaccharide analysis cannot provide information about the structures surrounding each disaccharide, but it is expected that differences between isoforms in 3S component production ratios reflect differences in specificity for surrounding structures.
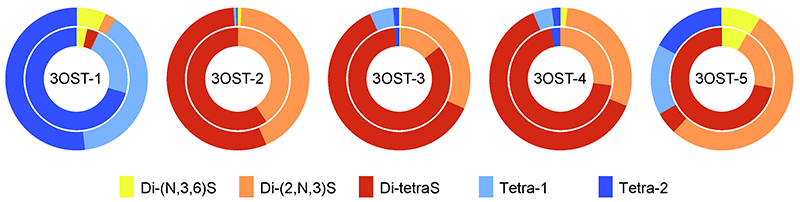
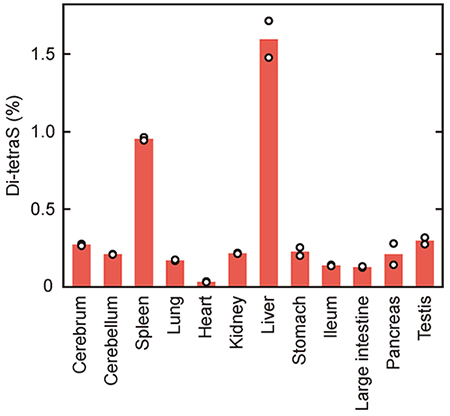
We for the first time identified the Di-tetraS unit, with all four sulfation loci modified, as a reaction product of 3OST-511. No information about the Di-tetraS unit was previously available, and it was unknown whether this novel structure is simply a by-product of 3OST-5, or has a physiological function of some sort. In order to explore these possibilities, the reaction specificities of other isoforms were investigated, and the Di-tetraS unit was found to be the principal reaction product for multiple isoforms (Fig. 6). Next, in order to ascertain the in vivo presence of the Di-tetraS unit, disaccharide analysis of HS from rat tissues was performed, and minute quantities of Di-tetraS were detected in all tissues examined (Fig. 7). The content was only approximately 0.2% of the total disaccharides, but this finding does show that the Di-tetraS unit is ubiquitous as an HS constituent. The Di-tetraS content was found to be especially high in the liver and spleen, and is expected to have specific roles in these organs.
In research on 3S, good progress is being made only with regard to AT-binding structure, whereas very little information has been obtained about non-AT-binding 3S. In recent years, it has become feasible to comprehensively analyze gene expression levels that are associated with physiological changes, and there have been multiple reports of phenomena suggesting links with 3OST. For example, in the pineal gland, which controls the circadian rhythm in rats, 3OST-2 is expressed only during the daytime12. In mouse embryos, the morphogenesis of submandibular gland is coordinated by 3OST-3 expression13. Drosophila with 3OST-B expression blocked shows abnormalities of the Notch signaling pathway14. In Caenorhabditis, 3OST is associated with neurite branching15. Aberrant expressions of 3OST genes have been reported in various cancers, although both promotional and suppressive effects have been found16. It is expected that the number of reports about physiological phenomena linked to 3OST gene expression will continue to increase in the future. However, research into the structure-function relationships of 3S is still difficult to perform, with a major difficulty being that no general method enabling direct detection and quantification of 3S in HS has been established. In this context, we have tried to measure 3S by the disaccharide analysis method, with five types of previously identified 3S components as standards17.
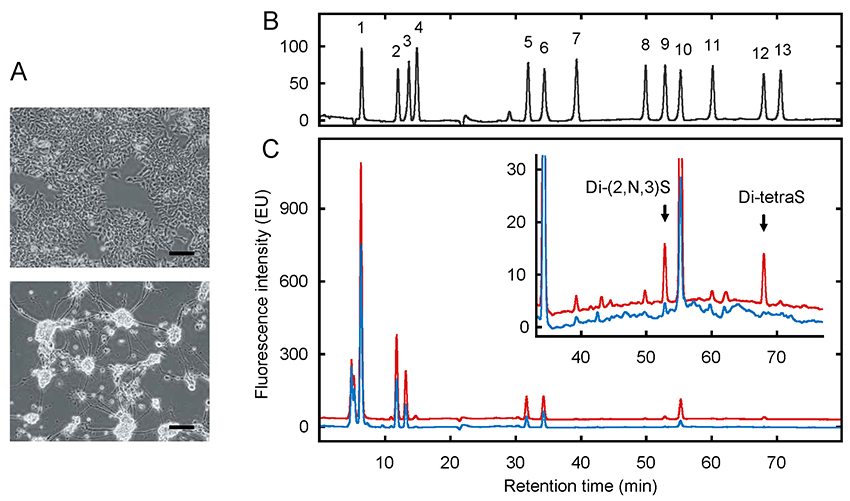
After sulfation with 3OST-5, HS and heparin include all five 3S components, and are therefore ideal raw materials for the preparation of standards. From the products of heparin lyase digestion of these, five components were separated and purified. Together with eight previously available disaccharide components, the total number of components was thus 13, and a mixture of these was prepared as a standard for quantitative analysis. Disaccharide analysis was performed using reverse-phase ion-pair HPLC with a post-column fluorescent labeling system. The method established by Toyoda et al. is both highly sensitive and highly specific, making it appropriate for measuring trace components such as 3S18. Optimizing the HPLC separation conditions enables baseline separation of all 13 components (Fig. 8B). In order to validate this analysis method, the HS from P19, mouse embryonic carcinoma cells, was analyzed. P19 cells have stem cell-like properties, and differentiate into neurons when stimulated by retinoic acid (Fig. 8A). As shown in Fig. 5, specific 3OST isoforms are expressed in the central nervous system, and it is therefore expected that changes in 3S associated with neuronal differentiation will be detected. The analysis results (Fig. 8C) were that, whereas almost no 3S was detected in undifferentiated P19 cells (blue chromatogram), after differentiation, Di-(2,N,3)S and Di-tetraS were detected in the neurons (red chromatogram), each making up 0.3% of the disaccharide content. The quantity of sample needed for a single analysis was 20 mg of cell acetone powder, which is equivalent to the amount prepared from two T75 flasks. HS from rat tissues was also analyzed, and measurement was feasible with the same quantity of sample. Therefore, use of this method enables quantitative detection of 3S in various biological samples.
More than 1,000 reports have been published about functional proteins that bind to HS or have physiological functions coordinated by HS or both. On the other hand, it cannot be denied that there is a shortage of information about the structure-function relationships of HS. One reason for this is that no effective methods exist for analyzing HS, with its complex and heterogeneous structure, but another reason is that the progress of research relating to 3S has been slow. In most previous studies, 3S was not examined, so not even its possible existence was considered. The vagueness of structure-function relationships may thus result from ignorance about 3S, the key to elucidating the structure-function relationship. The author therefore considers it essential to verify again the possible involvement of 3S in known physiological activities of HS.