
Motomitsu Kitaoka
Faculty of Agriculture, Niigata University. Ph.D., Agriculture.
Graduated from Faculty of Engineering, The University of Tokyo in 1985, and after working for a private company, he earned his Ph. D. degree in 1993. From 1995 to 1998, he studied the reaction mechanism of dextran synthase as a postdoctoral fellow at Iowa State University. From 1998 to 2019, he was engaged in research focusing on the development of practical production technology for oligosaccharides using enzymes at the Food Research Institute, National Agriculture and Food Research Organization. He is now a professor at Niigata University since 2019.

Takane Katayama
Professor, Graduate School of Biostudies, Kyoto University
Bachelor of Science in Agriculture, Kyoto University, in 1994. Master of Science in Agriculture, Kyoto University in 1996. Ph.D, Kyoto University, Research Associate at Graduate School of Agriculture, Kyoto University in 1999.
Assistant Professor at Graduate School of Biostudies, Kyoto University in 2002. Assistant Professor at Ishikawa Prefectural University in 2005. Professor at Ishikawa Prefectural University in 2013. Professor at Kyoto University in 2015.
Takane Katayama is a Professor at Kyoto University. He received his Ph.D in the field of applied molecular microbiology under the supervision of Prof. Hidehiko Kumagai. After spending three years as a postdoc in the same lab, he moved to the lab led by Prof. Kenji Yamamoto and was appointed an assistant professor. In Prof. Yamamoto’s lab, he isolated the genes for 1,2-α-L-fucosidase and endo-α-N-acetylgalactosamindase from bifidobacteria, both of which are enzymes acting on human-derived glycans. These findings prompted him to consider host glycans-mediated symbiosis between gut microbes and humans. In the last decade, he has focused on functional analysis of bifidobacterial genes and enzymes involved in degradation of human milk oligosaccharides. His research has significantly contributed to revealing how bifidobacteria-rich microbiota is formed in the gut of breast-fed infants.
In general, a bifidobacteria-predominant microbiota is established in breastfed infant guts, and such microbiota formation is believed to be beneficial to host health. Since the isolation of Bifidobacterium sp. in 1899, research has been conducted to elucidate how bifidobacteria proliferate in the infant gut and which components, if present, in breastmilk promote their growth. In the 1950s, human milk oligosaccharides (HMOs) were reported to act as a growth factor for bifidobacteria. However, until relatively recently, the structural complexity of HMOs has hampered the elucidation of the molecular mechanism underlying HMOs-mediated selective growth of bifidobacteria. In the early 21st century, systematic understanding of the HMO utilization mechanism of bifidobacteria rapidly advanced. In this chapter, we describe the historical aspects and recent progress of the research on HMOs and bifidobacteria.
“Bifidobacteria” is the generic term for strains belonging to the genus “Bifidobacterium”. Bifidobacteria were first isolated by Henri Tissier in 1899 from the feces of a breastfed infant. At that time in Europe, bottle-fed infants often suffered from infectious diarrhea. Tissier observed that bifidobacteria were seldom found in the feces of bottle-fed infants while they were frequently found in the feces of breastfed infants. He thus hypothesized that a bifidobacteria-dominant intestinal microbiota contributes to the prevention of diarrhea, thereby maintaining infant health1. It should be noted that nowadays, formula milk is supplemented with prebiotic*1 oligosaccharides to promote bifidobacterial growth, and the gut microbiota composition of bottle-fed infants is comparable to that of breastfed infants2,3. Tissier recorded the strain as “Bacillus bifidus communis” and proposed “Bifidobacterium” as a new genus name. However, the name was not officially adopted until 75 years after the proposal (Bergey's Manual of Determinative Bacteriology, 8th edition, 1974).
*1Prebiotics: A substrate that is selectively utilized by host microorganisms conferring a health benefit4.Given the above background, human milk was expected to contain a “bifidus factor”, a component(s) that selectively proliferates bifidobacteria. Studies to isolate and identify the bifidus factor began around 1910, and in 1953, György and colleagues reported that, by examining the growth-promoting ability of fractionated human milk extracts on Bifidobacterium bifidum var. Pennsylvanicus (classified as Lactobacillus bifidus at the time), the bifidus factor could be oligosaccharides consisting of N-acetylglucosamine (GlcNAc), fucose (Fuc), galactose (Gal), and glucose (Glc)5. Since then, human milk oligosaccharides (HMOs), a mixture of oligosaccharides with a degree of polymerization of ≥ 3, have been assumed to be the bifidus factor in human milk.
An overview of HMOs appears in the first article in this series6. The most characteristic feature of HMOs is a richness of lacto-N-tetraose (Galβ3GlcNAcβ3Galβ4Glc, LNT) with a type-1 chain as the core structure. As such, non-human primate milk either does not contain LNT or only contains a small amount if present7.
Until relatively recently, due to the structural and compositional complexity of HMOs, it had been difficult to systematically understand how bifidobacteria decompose and utilize HMOs. In 1999, Bouquelet and colleagues identified 1,3-β-galactosyl-N-acetylhexosamine phosphorylase (GLNBP) that specifically phosphorolizes lacto-N-biose I (LNB, Galβ3GlcNAc), a disaccharide structure contained in LNT that represents the type-1 chain, and galacto-N-biose (Galβ3GalNAc), a disaccharide contained in mucin glycans, from a cell-free extract of Bifidobacterium bifidum8. Kitaoka et al. subsequently cloned the gene from Bifidobacterium longum subsp. longum (B. longum) and found that the homologs were found in multiple Bifidobacterium species. The findings prompted them to consider the physiological role of GLNBP of Bifidobacterium species residing in human guts9,10. They found that Bifidobacterium breve, Bifidobacterium longum subsp. infantis (B. infantis), B. longum, and B. bifidum, which are four species often isolated from infant feces, possessed the gene, and were able to ferment LNB. By contrast, Bifidobacterium adolescentis and Bifidobacterium catenulatum, which are often isolated from adult feces, did not have the gene, and were not able to utilize LNB11. Based on these results, Kitaoka et al. have proposed the “lacto-N-biose I hypothesis”, which differentiates infant gut-associated Bifidobacterium species from other species. They also predicted the HMO-utilization pathway of such Bifidobacterium species, which again provided evidence supporting the notion that HMOs serve as the bifidus factor9.
Based on the lacto-N-biose I hypothesis, a series of enzymes involved in the degradation of HMOs were identified and characterized. As a result, all glycosidases required for digesting the linkages of HMOs and some of the transporters necessary for importing HMOs have been identified to date (See chapter 4 for details). An interesting coincidence is that David A. Mills and colleagues were also aware of a high HMO utilization capability in B. infantis. They subsequently determined the genomic sequence of the type strain ATCC15697 (= JCM 1222) to understand the molecular basis underlying its HMO metabolism12.
We depicted the HMO assimilation pathways of the four major infant gut-associated bifidobacterial species, i.e., B. breve, B. infantis, B. longum, and B. bifidum in Figure 1. The functions of the respective enzymes are described later, but it is noteworthy that bifidobacteria employ two different HMO degradation pathways, depending on species/strains. One is a cell wall-anchored enzyme-dependent type that decomposes HMOs into mono- or di-saccharides extracellularly, and another is a transport-dependent one that internalizes HMOs in intact forms.
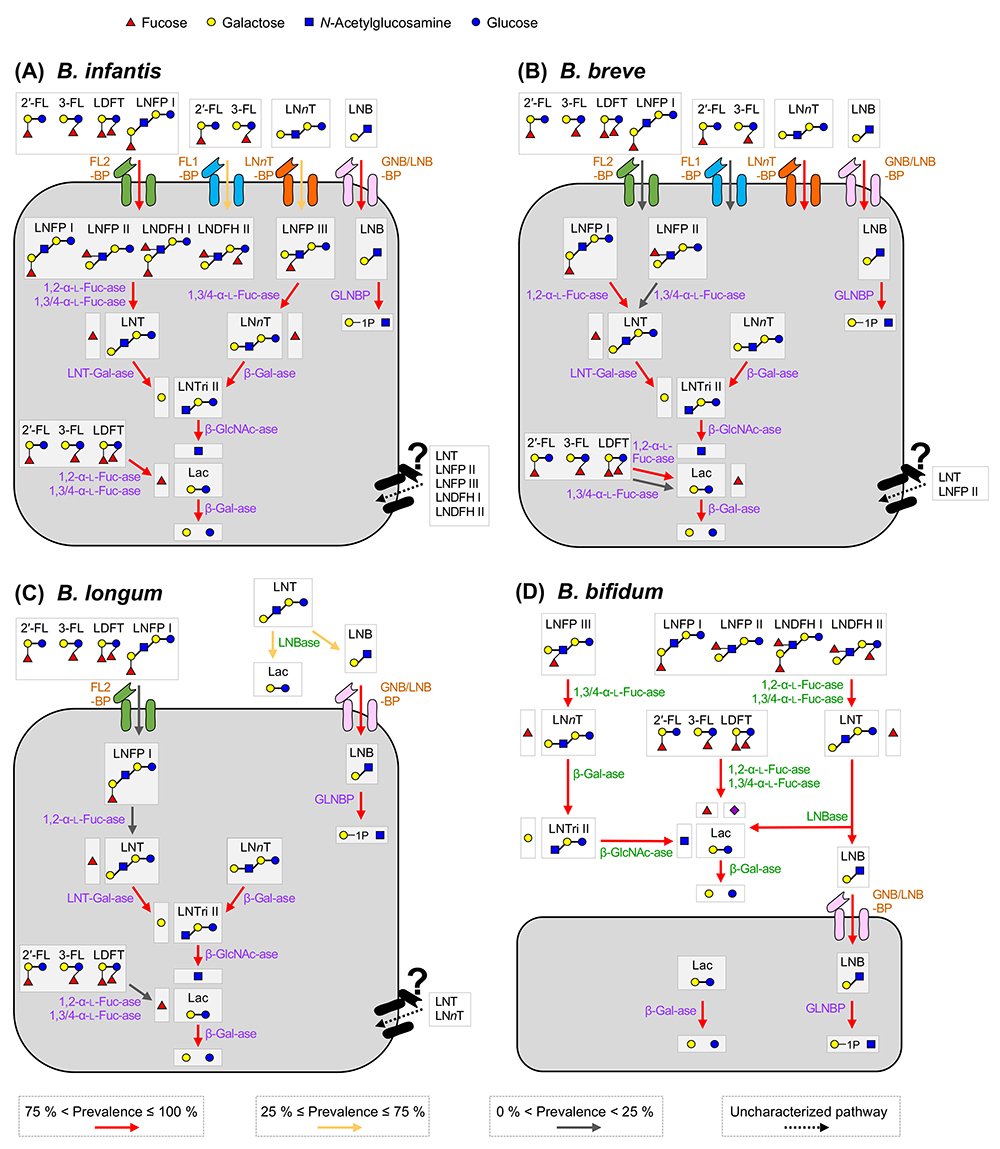
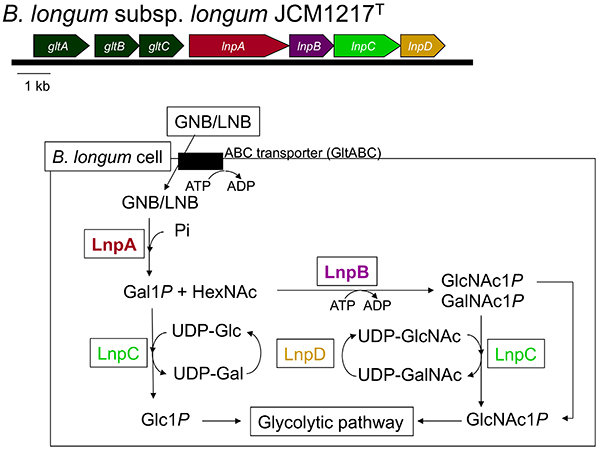
The above-mentioned 1,3-β-galactosyl-N-acetylhexosamine phosphorylase (GLNBP) gene forms a gene cluster with several other genes necessary for GNB and LNB assimilation (GNB/LNB pathway) (Figure 2). The pathway consists of GLNBP, the specific transporter for GNB and LNB13, and the enzymes that convert the GLNBP reaction products into the substances that can undergo the glycolytic pathway14. In general, the cluster consists of 7 genes (gltABC-lnpABCD), though some structural variations in the genomes are seen depending on the species.
gltABC encodes an ATP-binding cassette (ABC) transporter. GltA is a substrate-binding protein that specifically captures GNB and LNB, but not LacNAc. It is also capable of binding to LNT; however, the transporter is not considered to import LNT as the disruption of the extracellular lacto-N-biosidase gene (Fig.1C; lnbX, described below) of B. longum JCM 31944 markedly reduced the growth of the strain on LNT15.
lnpA encodes the above-mentioned GLNBP that exclusively phosphorolizes GNB and LNB to produce galactose 1-phosphate (Gal1P) and the corresponding N-acetylhexosamine. Compared to hydrolysis, phosphorolysis uses one less molecule of ATP before entering glycolysis. lnpB encodes N-acetylhexosamine 1-kinase (NahK), which phosphorylates α-anomeric hydroxyl groups of GlcNAc and GalNAc. lnpC encodes an enzyme (GalT) that transfers the UMP group from UDP-Glc/Gal to Gal1P/Glc1P, and lnpD encodes an enzyme (GalE) that reversibly converts UDP-Gal into UDP-Glc. GalET are enzymes that constitute a part of the Leloir pathway, a major galactose metabolizing pathway, by which Gal1P is sent to the glycolytic pathway via conversion into Glc1P. Notably, unlike the typical GalET of the Leloir pathway, LnpCD also acts on GlcNAc/GalNAc. As a result, the GalNAc1P generated by LnpB (NahK) is converted to GlcNAc1P to undergo the glycolytic pathway.
In summary, the GNB/LNB pathway is a route for importing GNB/LNB into the cytosol to be later subjected to glycolysis. The GNB/LNB pathway was present in all strains of infant-type B. breve, B. infantis, B. longum, and B. bifidum, but absent in adult-type B. adolescentis and B. catenulatum, as well as Bifidobacterium animalis subsp. lactis11.
To assimilate LNT via the GNB/LNB pathway, LNT must be degraded by lacto-N-biosidase (LNBase) into LNB and lactose (Lac). When the LNB hypothesis was initially proposed, only one LNBase belonging to glycoside hydrolase family (GH) 20 was isolated from Streptomyces sp16. Within the Bifidobacterium genus, LNBase activity was detected in B. bifidum and some strains of B. longum. Subsequently, the GH20 LNBase gene (lnbB) was isolated from B. bifidum, while a novel LNBase (lnbX) was isolated from B. longum, which was then classified into a new family GH13617,18. The occurrence of LNBase in Bifidobacterium is limited as lnbB is found only in B. bifidum and lnbX is found in B. bifidum and approximately half strains of B. longum. All of these enzymes are cell wall-anchored enzymes.
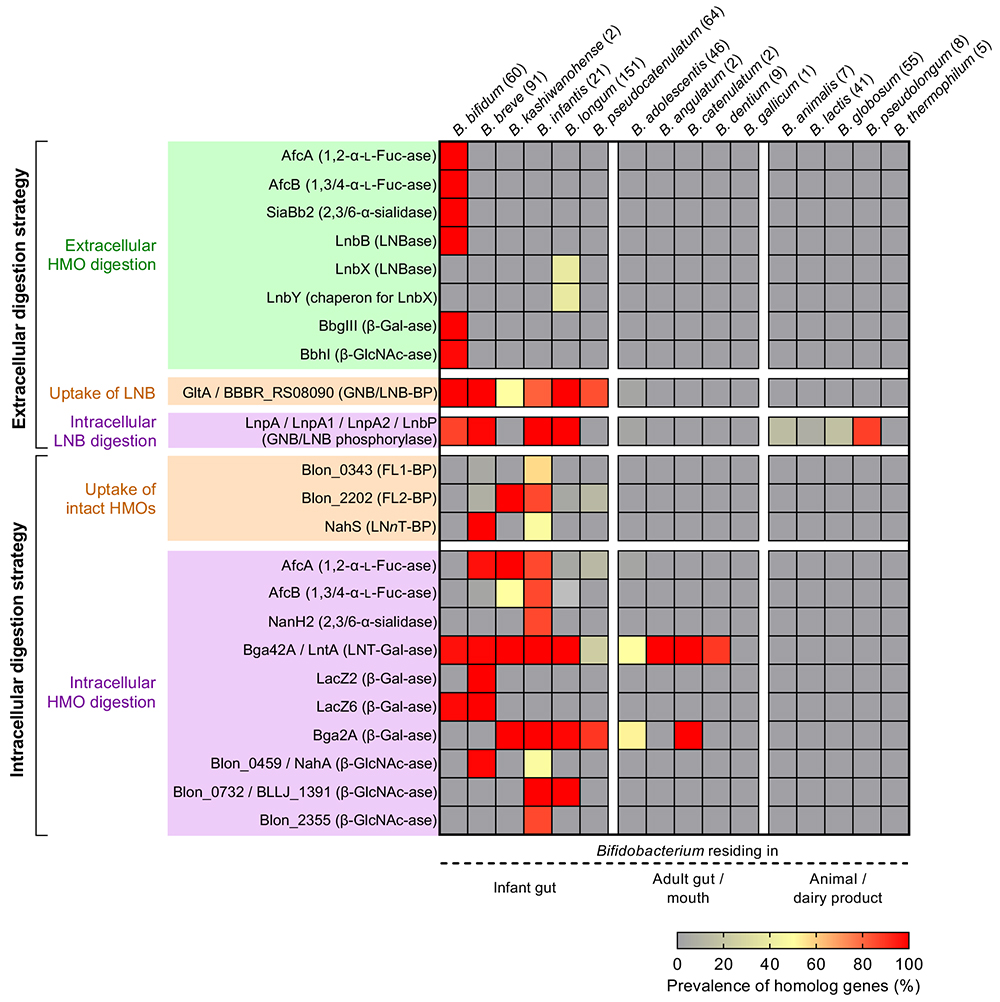
HMOs are comprised of 20 core structures that are frequently modified by fucose and/or sialic acid residues19. 1,2-α-Fucosidase belonging to GH9520 and 1,3/4-α-fucosidase belonging to GH2921 have been identified as the enzymes that specifically liberate fucose from H- and Lewis-antigen structures, respectively. α-Sialidase belonging to GH33 removes α-(2→3/6)-linked sialic acid residues22. The distributions of these enzymes in Bifidobacterium species are shown in Figure 3. In B. bifidum, these enzymes are anchored to cell walls, while in other species they are localized at cytoplasmic space.
As described in the next chapter, B. infantis imports all HMOs as intact forms. B. infantis has the GNB/LNB pathway but it lacks LNBase; therefore, the degradation pathway of LNT in this species remained to be elucidated. Yoshida et al. isolated a cytosolic β-galactosidase belonging to GH42 that showed a high activity on LNT (LNT-1, 3-β-galactosidase) from B. infantis JCM 122223. Cytosolic GH20 β-N-acetylglucosaminidases with varied similarities were also isolated from B. infantis and B. longum24,25. GH42 β-galactosidase homologs are distributed not only in infant gut-associated species but also in some adult gut-associated species of Bifidobacterium, while GH20 cytosolic β-N-acetylglucosaminidase homologs are distributed among infant gut-associated species. LNT internalized into the cytoplasmic space is therefore sequentially hydrolyzed into monosaccharides from the non-reducing end by these glycosidases, and not utilized via the GNB/LNB pathway.
In 2011, Asakuma and colleagues characterized the HMO utilization patterns of the four major infant-gut associated Bifidobacterium species to functionally link the phenotypes with the genotypes of those strains26. They cultured B. bifidum JCM 1254, B. longum JCM 1217, B. infantis JCM 1222, and B. breve JCM 1192 in a medium supplemented with neutral HMO fractions purified from human milk as a sole carbon source, and quantified each HMO and the degraded products in the spent media. B. bifidum and B. infantis exhibited vigorous growth in HMO-medium, but the growth of B. longum and B. breve was poor. In the culture supernatant of B. bifidum, which possesses many cell-wall-anchored HMO degrading enzymes, HMOs were first decomposed into mono- and disaccharides and then utilized by the cells. Contrary, no decomposition was detected in the culture supernatant of B. infantis (a slight increase in monosaccharide concentrations was temporarily detected, which was considered to be a counteraction against an osmotic pressure), indicating the direct internalization of HMOs. B. bifidum degraded LNT and lacto-N-fucopentaose I (LNFP I: Fucα2Galβ3GlcNAcβ3Galβ4Glc) during the early phases of the cultivation, into Fuc, LNB, Lac, and Gal. LNB and Lac were consumed immediately, but Fuc and Gal remained unconsumed. The results suggested that the HMO degradation products may be cross-fed to other bacteria. Gotoh and colleagues then demonstrated the altruistic properties of B. bifidum by culturing human fecal suspensions in medium supplemented with HMOs. When B. bifidum was added to the culture, bifidobacterial species other than B. bifidum proliferated and the abundance of Bifidobacterium within the entire microbiota significantly increased. The growth-stimulatory effect of exogenously added B. bifidum was not observed when Glc was used as a carbon source, and even in an HMO-added medium, the effect disappeared in the presence of a fucosidase inhibitor27. Interestingly, prior to this report, Tannock and colleagues showed that the occupancy of Bifidobacterium in the microbiota is high in the infants’ feces when the B. bifidum is present > 10 % in the bifidobacterial community. They reported that that the tendency was observed only in breastfed infants and was not found in infants fed with formula prepared from cow milk or goat milk, which contain almost no oligosaccharides other than lactose28.
B. longum and B. breve consumed LNT only. A transient increase in LNB, which is produced by the cell-wall-anchored LnbX (GH136 LNBase), was observed in the culture supernatant of B. longum. Further research revealed that LnbX-positive B. longum strains also cross-feed the degraded products to other species (LNB and/or Lac)15. As mentioned above, in the culture supernatant of B. infantis, all HMOs decreased at a similar rate, indicating the selfish behavior of this subspecies. It should be noted that the characterization of transporters is still lagging, as only three HMO transporters have been physiologically identified.
The prevalence of glycosidases, phosphorylases, and ABC transporters responsible for the utilization of HMOs in the genus Bifidobacterium was systematically analyzed using publicly available genomic sequences (Figure 3)29. The genes are widely distributed in the infant gut-associated species of Bifidobacterium with minor exceptions, suggesting the co-evolution between humans and Bifidobacterium mediated through HMOs.
Several other gut microbes belonging to genus Bacteroides and phylum Firmicutes, and Akkermansia muciniphila are also known to assimilate HMOs, but the efficacy is much less than that of B. bifidum and B. infantis30,31. Interestingly, butyrate-producing species belonging to genus Roseburia in phylum Firmicutes are reported to utilize not only a few HMO molecules but also plant-derived glycans such as xylooligosaccharides. This ability may reflect a nutrient shift of infants from breastmilk to solid foods during the weaning period32.
After approximately 50 years after the isolation of Bifidobacterium sp. from the feces of breastfed infants, the role of HMOs as the bifidus factor was proposed5. However, a few years later, the same group published a paper describing GlcNAc auxotrophy of the strain they used33. The report might be considered to be a correction of their conclusion that HMOs are the bifidus factor. It then took another 50 years to reconsider the relationship between HMOs and bifidobacteria. Surprisingly, however, after the role of HMOs as a bifidus factor was established, commercialization of 2'-fucosyllactose as an additive to formula milk began within 10 years in Europe and the United States (will be mentioned later in this series by Yanashima34). While the rapid development is very impressive, we need to be aware that the 2'-fucosyllactose utilization capability is limited to certain strains within the genus Bifidobacterium (will be mentioned later in this series by Sakanaka35). The distribution of genes responsible for the assimilation of prebiotics, including HMOs, in Bifidobacterium, is likely a conceptual counterpart to the distribution of virulence genes in pathogenic bacteria. Clinical applications of bifidobacteria as probiotics and HMOs as prebiotics should be implemented with a strain-level understanding of their genotypes and phenotypes36.