
Sachiko Akase
After graduating from Department of Bioinformatics at Soka University, I have been studying glycoinformatics with Dr. Kiyoko F. Aoki-Kinoshita as a master’s student in the Graduate School of Engineering. I perform simulation of N-glycan synthesis in Drosophila by utilizing the enzyme information registered in FlyGlycoDB.

Kiyohiko Angata
Graduated from the Institute of Biological Sciences at University of Tsukuba, Trained and learned glycobiology under the supervision of Dr. Minoru Fukuda at La Jolla Cancer Research Foundation (current Sanford Burnham Prebys Medical Discovery Institute). Research topics are profiling of glycogene expression, analysis and application of new function of glycans, and development of glyco-DB (ACGG-DB).
In the third article of this series on Glycan and Database, we introduce glycan-related gene databases. Genes associated with glycan synthesis (glycan-related genes, glycogenes) include genes necessary for glycosynthesis such as sugar-nucleotide synthases, sugar-nucleotide transporters, and glycosyltransferases, and genes necessary for glycolysis such as glycosidase. In humans, about 300 glycogenes are involved in the synthesis and breakdown of glycans. During the evolution of organisms, the number of glycogenes has increased and their kinds have become increasingly diverse. Here, we first introduce databases (DBs) useful for research on glycogenes, and then describe the features of our databases, i.e., GGDB and FlyGlycoDB, which provide information about human glycogenes and fly glycogenes, respectively.
Glycoscience research has so far been focused on the glycan structure and biosynthesis, but there are an increasing number of studies being carried out within many different disciplines that seek to elucidate the functions of glycans. These disciplines include microbiology, developmental biology, immunology and oncology. Under these circumstances, DBs which integrate a range of information about glycoscience research may be as useful as relevant literature for new researchers who have not previously studied glycans. When the name of a particular glycogene is found in a paper or appears in an analyzed result, general DBs or knowledge bases (KBs) for genes and proteins can be used as a starting point to find out more about the glycogene [in question]. The DBs on glycogenes introduced here are also publicly available as tools for collecting relevant information. Selecting the DB that is best suited to the research objective is the most efficient approach and takes advantage of the characteristics of individual DBs according to the objective.
For example, when you try to obtain information about the cDNA or genome structure and the sequence of a certain glycogene, you may want to utilize gene, Ensembl, and CAZy. In particular, CAZy covers the glycogenes of several species and is useful for comparing different species and performing evolutional analysis. OMIM (https://www.ncbi.nlm.nih.gov/omim, https://omim.org) and the Human Protein Atlas are useful for investigating relationships to disease. With the Human Protein Atlas, you can find the quantity of glycogene expression in various types of cancer cells. The databases used for protein analysis include KEGG, UniProtKB, GlyGen, and GeneCards. In these databases, you can access pathways, mass spectrometry and interactome analysis results.
For humans, about 300 glycogenes have been cloned so far. Among these, the GlycoGene Database (GGDB) focuses on the glycogenes involved in glycosynthesis and has been made publicly available as a Japan Consortium for Glycobiology and Glycotechnology DataBase (JCGGDB). At present, the most recent version, which reflects changes made to the display method, interface-related matters, and descriptions, is publicly available in ACGG site (Narimatsu et al. 2017). Here, we introduce its features.
*GGDB is accessible from not only the website of ACGG but also that of the Japan Consortium for Glycobiology and Glycotechnology (JCGG) and the GlyCosmos portal.
At present, 221 glycogenes involved in glycosynthesis are listed. On the top page, the glycogenes are displayed in alphabetical order, which makes it easy for you to find the enzyme reactions and key words you are interested in (Fig. 1).
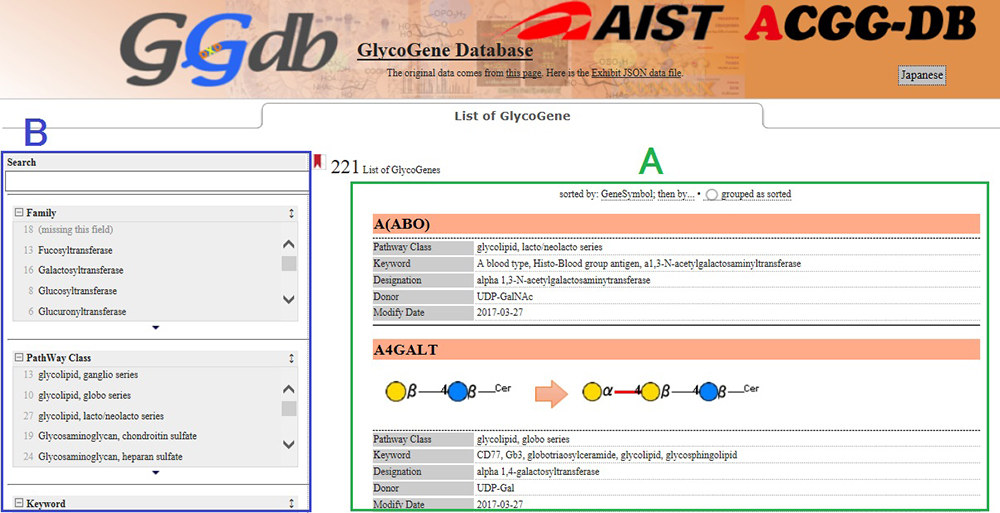
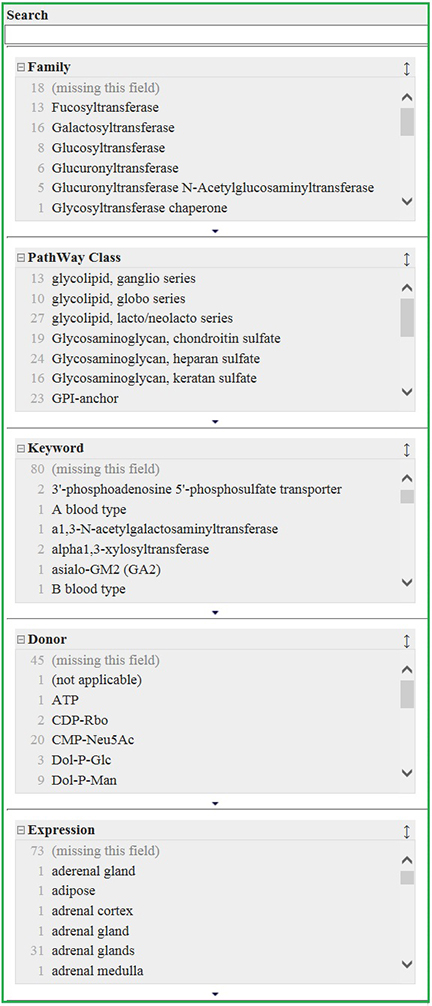
As search modes, Text Search and Facet Search are available, which makes it easy for you to search for the particular glycogene you are interested in (Fig. 2).
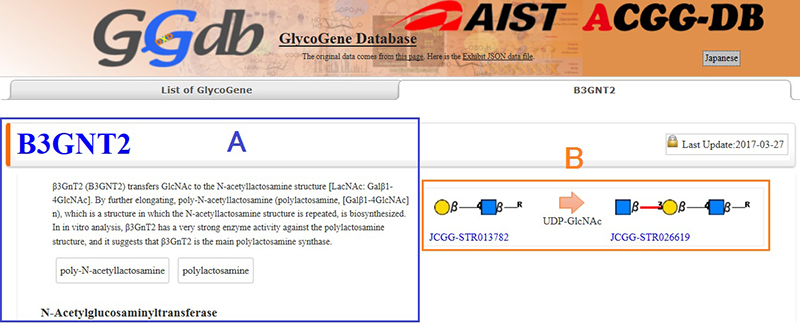
Next, we explain what can be found on the page for a selected glycogene.
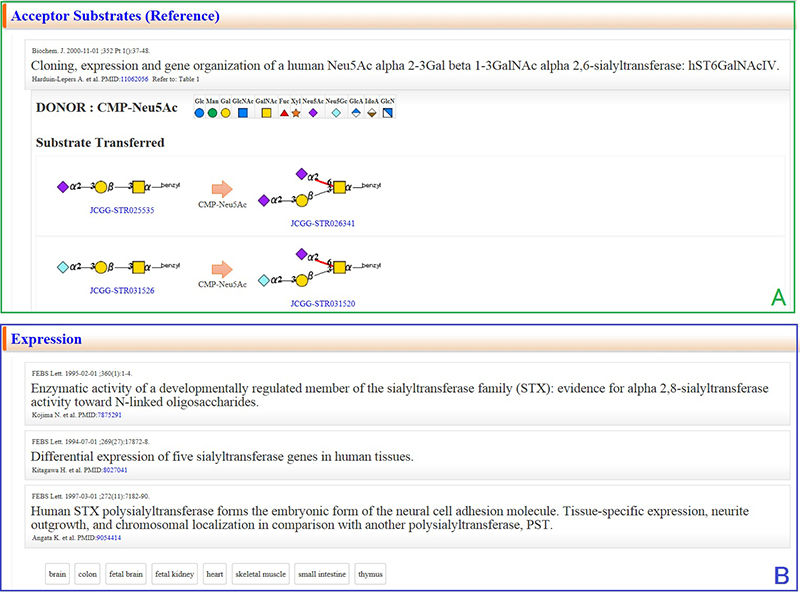
On the page for the selected glycogene, a summary description of the glycogene is provided and the main enzyme reaction is shown with precursor, substrate (sugar nucleotide), and reaction products (Fig. 3). The feature common to ACGG-DB including GGDB is that all databases use a common language, i.e., glycan structure, (JCGG-STR). The above general DBs provide information using words alone, whereas ACGG-DB enables researchers to intuitively imagine reaction products: this is one of the unique features of ACGG-DB. For the summary section, glycoscience researchers at the National Institute of Advanced Industrial Science and Technology (AIST) are responsible for its curation: the enzyme reactions and functions of the gene are briefly summarized.
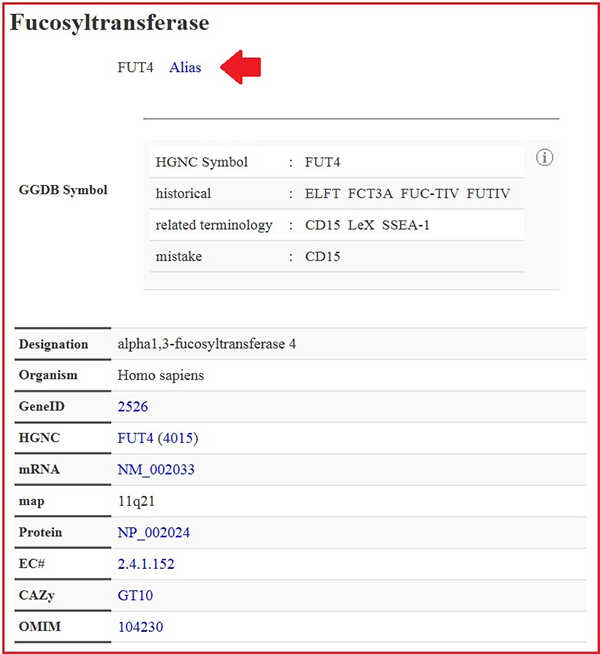
The gene names provided by HGNC are used. The common names that have been historically used are stored in Alias. Over the long history of glycoscience, the same glycogenes have been given different names as synonyms. For example, the gene FUT4 has been referred to as the gene CD15 because CD15 is a product of FUT4, and CD15 is stored as a synonym in Alias. However, in order to avoid confusion, the term CD15 should no longer be used and is provided in Alias just for reference (Fig. 4).
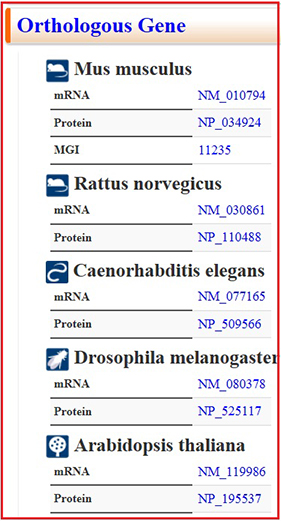
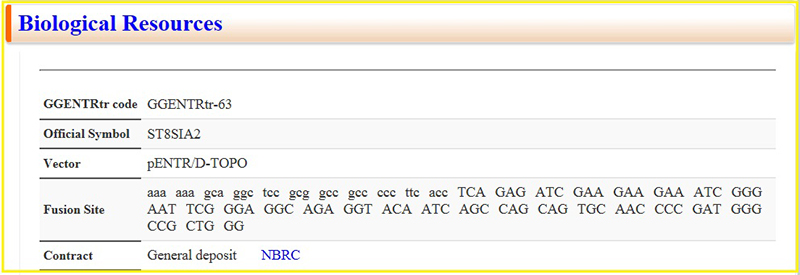
The mid and lower sections provide information about homologous genes (Fig. 5), enzyme reactions and mRNA expression (Fig. 6), and resources (Fig. 7).
Even in 2019, new glycogenes as well as new activities and functions of glycogenes are being reported, which means that summaries and enzyme reactions need to be updated periodically. Information about mutations and expression quantities of glycogenes associated with glycan synthesis pathways may be useful for research on diseases or carcinomas with accompanying glycan abnormalities. Going forward, we intend to promote collaboration within GlyCosmos in order to develop a method of utilizing GlyCosmos in conjunction with the other DBs in a cross-cutting manner.
FlyGlycoDB is a database for glycan-related genes in Drosophila melanogaster (scientific name, hereinafter to be referred to as "Drosophila") and is available at http://fly.glycoinfo.org/. Drosophila has long been used as a model organism in not only glycoscience research but also many other fields of biological research. FlyBase (Thurmond et al. 2019) is a database which summarizes genetic study results on Drosophila and stores all the recorded genes of Drosophila. It is difficult, however, to extract glycan-related genes from FlyBase. No databases have focused solely on the glycan-related genes of Drosophila. Under these circumstances, FlyGlycoDB was developed as a database which can be easily utilized in glycan research. In FlyGlycoDB, a total of 160 glycan-related Drosophila genes for which functions have been identified are currently registered, including genes for which activity has been confirmed by analysis using RNAi silencing (Nishihara et al. 2007, 2010; Yamamoto et al. 2015) and genes obtained from CAZy (Lombard et al. 2014), which is a database for carbohydrate-active enzymes. Fig. 8 shows the top page of FlyGlycoDB and Fig. 9, the entry page for each gene.
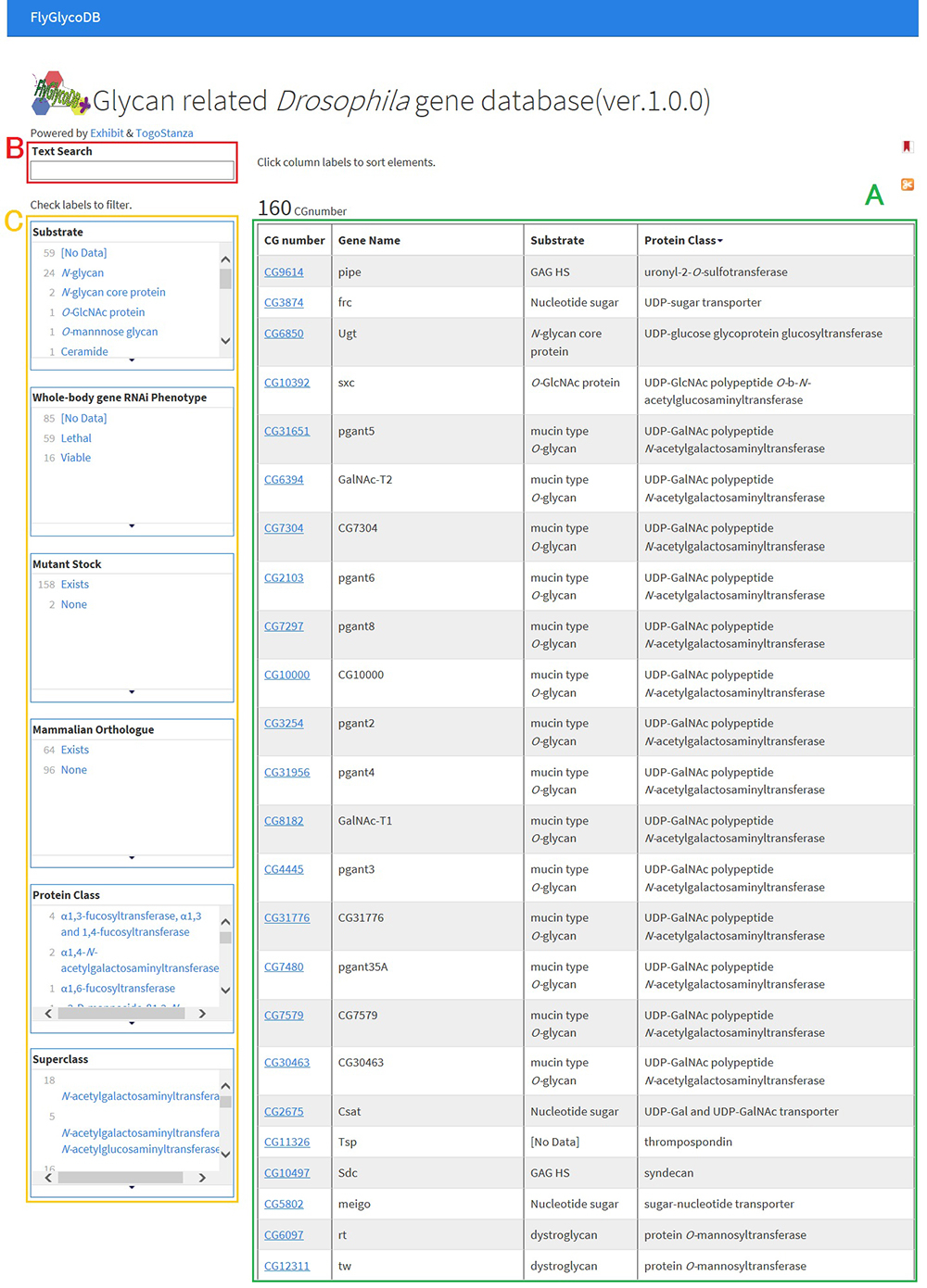
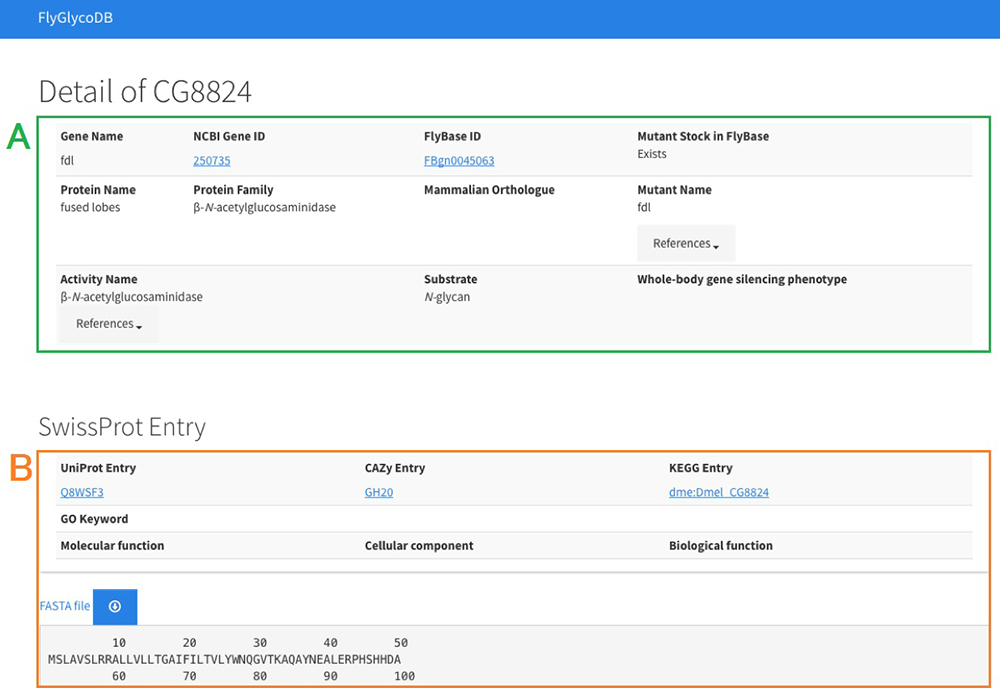
FlyGlycoDB is available through Semantic Web services and can easily be used in conjunction with external databases. Therefore, the entry page for the selected gene provides not only the information about that gene that is stored in FlyGlycoDB but also the information obtained from SwissProt.
FlyGlycoDB will be updated and made available from GlyCosmos as of August 1, 2019: a total of 166 entries, can be browsed according to gene classification, will be available under GlyCosmos Data Resources.