Issaku Yamada
Received his Ph.D in engineering from Tokyo Metropolitan University in 1997. After having worked at Nagoya University (fellowship from Japan Society for Promotion of Science [COE program]), he became a Noguchi Institute researcher in 2002 and started scientific investigation of glycans at the Institute in 2006. He is presently engaging in glycan informatics studies on research topics such as glycan structure notation, ontology, and database design.

Daisuke Shinmachi
Received his master’s degree from Soka University, Department of Engineering. He joined the JST integration promotion program as an assistant researcher at Soka University in 2014. He is currently working at SparqLite and is developing a system of glycan informatics using semantic web technology.

Masaaki Shiota
Received his master’s degree from Soka University Department of Engineering in 2017. He started working for the JST integration promotion program as an assistant researcher at Soka University in 2017 and is involved mainly in the development and operation of the GlyCosmos Portal as an engineer

Nobuaki Miura
Received his Ph.D in theoretical chemistry from Hokkaido University in 1998. After having worked as a researcher at several institutes, he became a specially appointed associate professor of Hokkaido University from 2003 to 2012 (responsible for Sun Microsystems donation Laboratory of Molecular Life Science from 2003 to 2008) and involved in glycan research. He started development of Total Glycome analysis software (TAG) (under development) while he worked at Ochanomizu University from 2013 to 2017. He is currently researching bioinformatics mainly of metaproteome using artificial intelligence as a specially appointed associate professor at Niigata University.

Shujiro Okuda
Received his Ph.D. from the Graduate School of Science, Kyoto University in 2007. After having worked at Bioinformatics Center, Institute for Chemical Research, Kyoto University as a postdoctoral researcher, he became an associate professor in the Biological Information Department, Life Science Division, Ritsumeikan University and started development of a database dedicated to the life science field and microbiome studies. He became an associate professor in the Department of Medical and Dental Science, Niigata University in 2013 and is currently researching bioinformatics in medicine.

Kiyoko F. Aoki-Kinoshita
Received her Ph.D. in computer engineering from Northwestern University in 1999. After a brief period as a post-doctoral fellow at the Institute of Information Science, Academia Sinica in Taiwan, she worked as a senior software engineer at BioDiscovery, Inc. in Los Angeles for three years. From 2006, she moved to the Bioinformatics Center, Institute of Chemical Research, Kyoto University, where she started her research career in glycoinformatics. She is now a professor at Soka University, where she currently teaches and continues to do research to develop useful glycoinformatics tools for the community and to apply them to furthering the understanding of glycan function in biological systems.
In the fifth installment of this series, glycan databases will be described. Glycans with various structures exist in the body and they also take the form of free glycans and complex carbohydrates such as glycoproteins and glycolipids. The glycan databases described below include: the international glycan structure repository GlyTouCan for glycan structures; GlyCosmos Glycans for glycan structures; GlycomeAtlas for visualizing localization of glycomes; TotalGlycome for analyzing glycan expression data; and GlycoEpitope for carbohydrate antigens and antibodies.
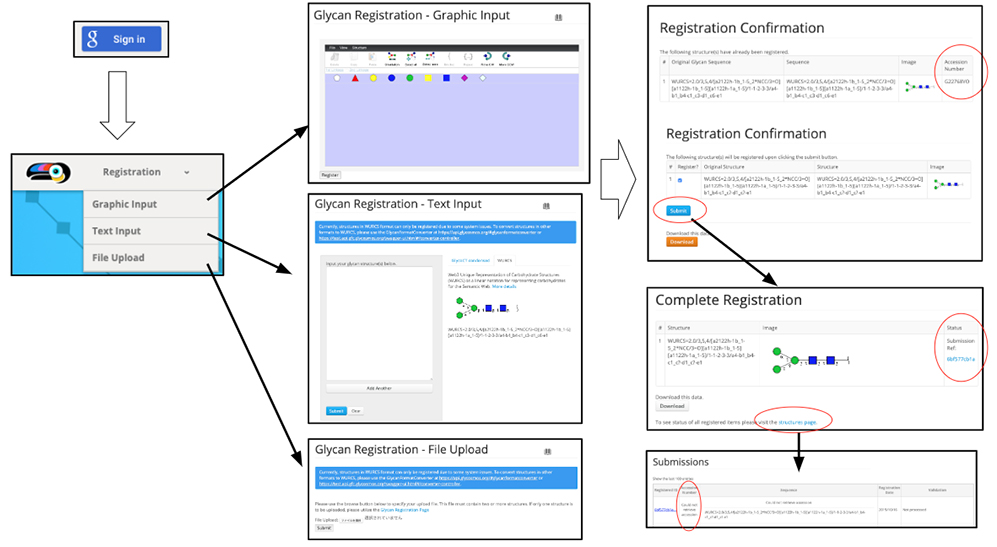
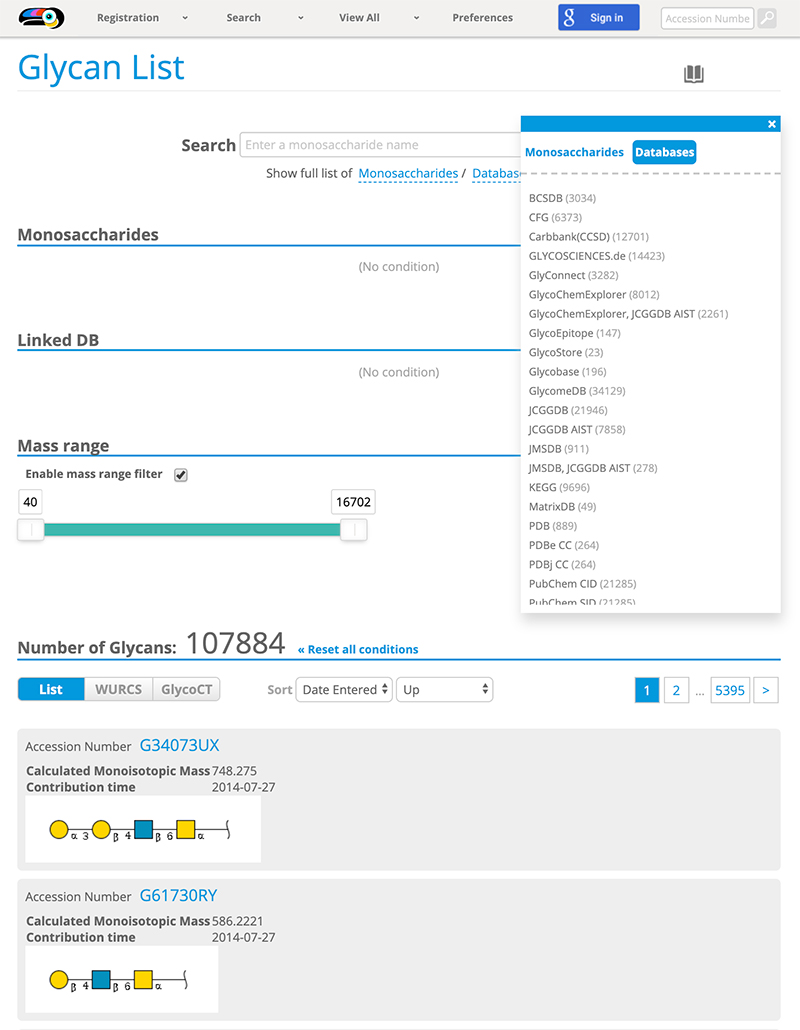
The necessity for clarifying glycan structures in scientific research was discussed at the ACGG-DB meeting convened in Dalian, China, in 2013. During the discussion, it was agreed that a repository of glycan structures would be constructed and a unique accession number would be assigned to each glycan structure 1. The international glycan structure repository called GlyTouCan 2 was released in August 2014 so that unique accession numbers could be assigned to each glycan structure. Initially, GlyTouCan was constructed based on the data in GlycomeDB 3 such as glycan structures, biological species, and links to other glycan databases. It currently stores information on a total of 115,802 glycan structures (as of October 3, 2019). GlyTouCan can be used free of charge on the internet (https://glytoucan.org). A user who wants to register a glycan structure will first “Sign in,” and then use one of three methods for registering a glycan structure from the “Registration” menu (Fig. 1). After the glycan structure is inputted and the “submit” button is clicked, the “Registration Confirmation” page is shown. If the glycan has already been registered, an accession number is shown on the right. No accession number means that the structure has not been registered. After the user inputs a structure and clicks the “Submit” button, the “Complete Registration” page is shown. The user then clicks “structures page” shown in blue at the bottom of the page, and a list of glycan structures contributed by the user will appear. The registration of these contributed glycan structures are processed by the GlyTouCan system on an hourly basis. After this in the “Submissions” page, the “Could not retrieve accession” shown in the “Accession Number” column (the second column of table in the page), is changed to an accession number starting with the letter “G,” which is an accession number assigned to the registered glycan structure. Then, the accession number of the registered glycan structure becomes available. To search for a registered glycan structure, either the “Search” bar or “View All” menu (Fig. 2) can be used. See “User Guide” for a detailed description of the procedure.
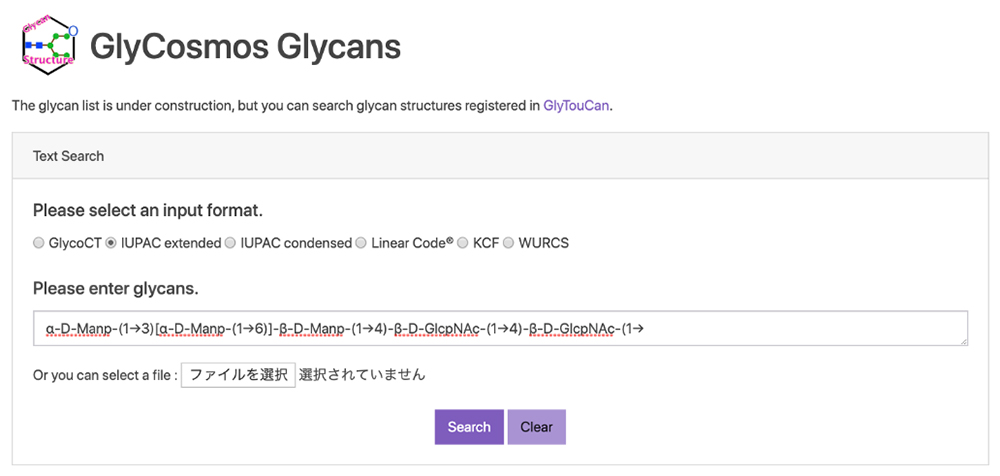
GlyCosmos Glycans was developed to make available the links to biological species, the literature, and external databases for glycans, because GlyTouCan was limited to the registration of glycan structures. A string can be used for search queries at present (as of October 16, 2019), where six types of glycan structure formats are available (GlycoCT 4, IUPAC extended 5, IUPAC condensed 5, Linear Code® 6, KCF 7, WURCS 8 ) (a href="#fig_3">Fig. 3). Major formats are described in the summary of glycan nomenclature and glycan-related resources in this series.
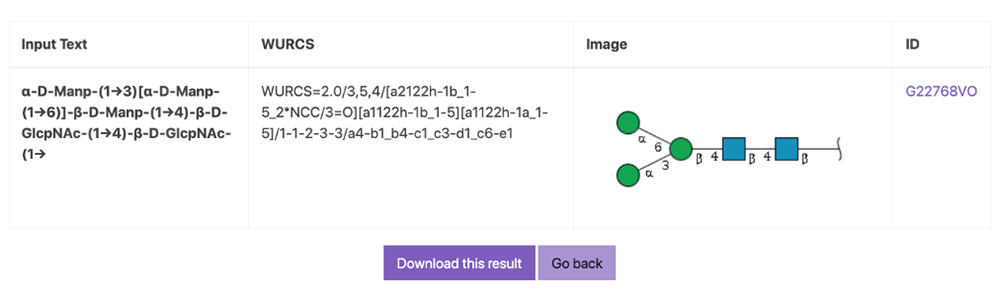
When the search result is “Not Found,” as indicated under ID on the right side of the page, the glycan structure of the search query has not been registered in the database (Fig. 4).

When the accession number starting with the letter “G” is shown, the glycan structure of the search query has been registered in the database (Fig. 5). When the user clicks the link here, the page containing the targeted glycan structure is shown. The entry page is linked to GlyTouCan at present. We are planning to develop more sophisticated datasets to provide information such as a list of species, lists of tissues and glycan structures by species, a list of monosaccharides, and links to other databases.
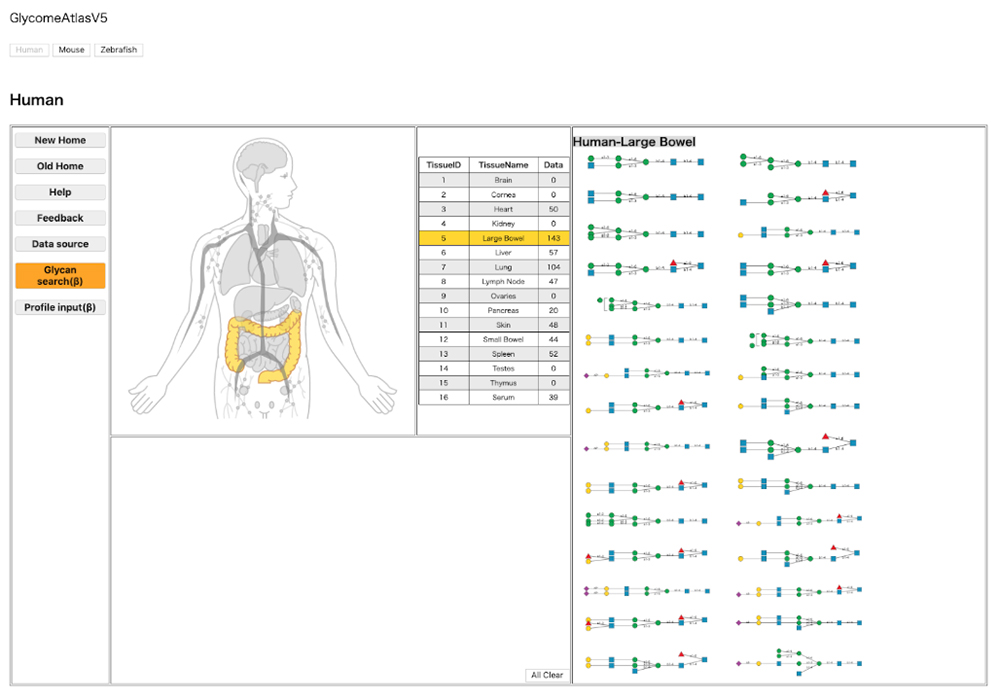
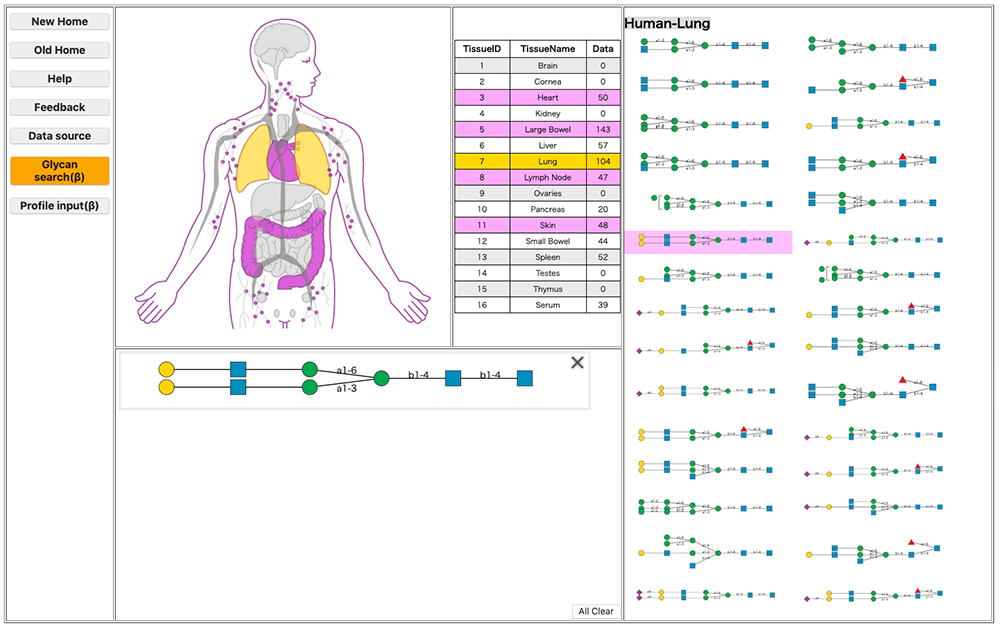
The structure of a glycan continuously changes in the body in association with the site of localization. Such changes in structure and site of localization are closely connected to the glycan’s function. Understanding the localization of a glycan is therefore vital to the scientific study of the glycan. GlycomeAtlas 9 enables visualization of glycan localization in the body. The localization of glycan structures can currently be visualized in humans, mice, and zebrafish (as of October 30, 2019). The source of glycan profiling data used for humans and mice is the CFG (Consortium for Functional Glycomics) , and that used for zebrafish has been published 10.
Switching species is possible by clicking the button “Human,” “Mouse,” or “Zebrafish” at the top of the page. When “Tissue Name” is clicked, the relevant tissue is highlighted and the glycan structure within is shown on the right-hand side of the page (Fig. 6). When a glycan structure shown on the right is selected, the site of localization of the glycan structure is highlighted (Fig. 7).
Glycans control an enormous amount of biological information and are involved in various biological processes such as intercellular communication in response to pathogens entering the body, immunity-related pathogen recognition, signal transduction, protein folding during biosynthesis, quality control of proteins, etc.
Change of glycan expression in various biological processes such as disease has been actively studied for a long time, since expression patterns of glycans change in response to such a process. In previous studies, for various reasons including measurement limitations, conventional research has mainly focused on one to several glycans (subglycomes). Recent progress in mass spectrometry and new analysis methods have enabled comprehensive analysis among different classes of glycans, with particular attention paid to the total glycome 11, 12. In total glycome analysis, qualitatively and quantitatively up to five classes of glycans can be analysed, namely, N-linked glycans (N-glycans), O-linked glycans (O-glycans), glycosphingolipid glycans (GSL-glycans), glycosaminoglycans (GAGs), and free oligosaccharides (fOSs). The method enables observation of the expression patterns of more than 200 glycans involved in biological events such as disease and differentiation. Investigation of the total glycome is important for the following reasons:
● There are various glycoconjugates on the cell surface, and the entire expression information can be used for marker search etc.
● A common epitope exists in various classes of glycans.
● Failure to synthesize a particular glycan repairs by another class of glycans.
● The metabolic pathways of N-glycans and N-type free oligosaccharides are closely related.
The Total Glycome Database is a part of the GlyCosmos portal and was constructed for the purpose of linking with other omics by observing Total Glycome comprehensively.
Total Glycome Database contains results of expression analyses of N-glycan, O-glycan, GSL-glycan, fOS, and GAG and provides various views to present data. A selected dataset is shown by choosing items from among the pulldown menus comprising “model”, “source”, “select a view”, and “glycan class” at the top of database screen and clicking the “Submit” button. Currently available options for viewing the glycan expression data include tables, bar charts for comparing glycan expressions, and pie charts for expression changes in the percentage of glycan expression in each class (Fig. 8).
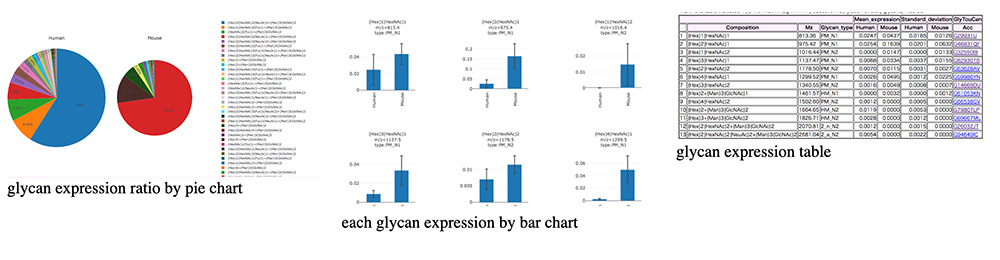
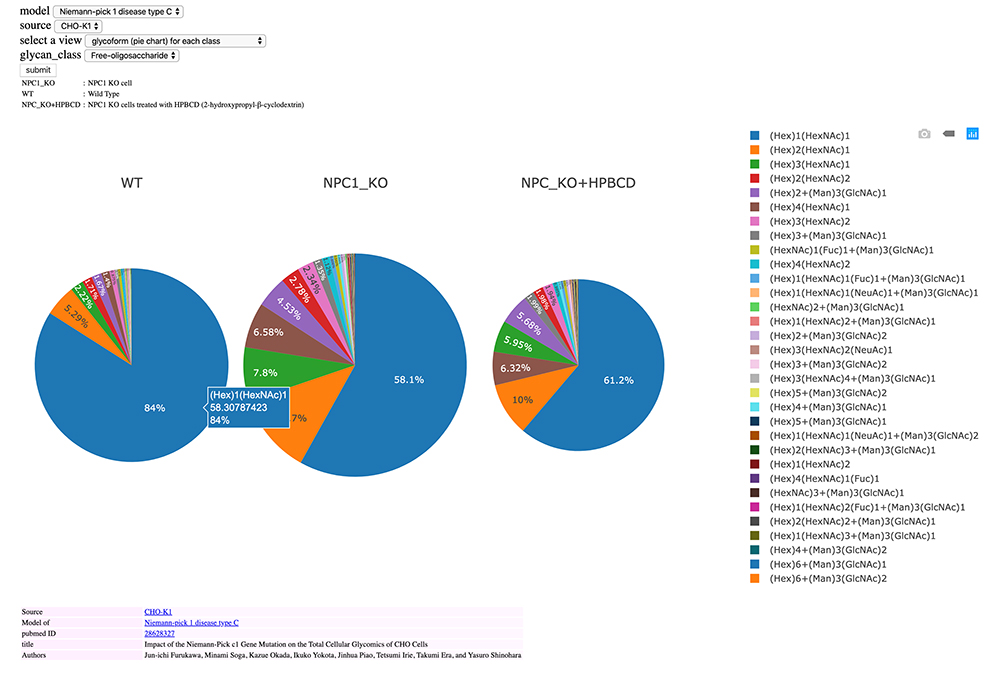
Fig. 9 shows glycan expression as a pie chart for three different cells: wild-type CHO cells (WT); CHO cells after knockout of the Niemann-Pick disease type C1 (NPC1) gene; and CHO cells treated with 2-hydroxypropyl-β-cyclodextrin (HPBCD), a drug candidate reported to have efficacy for NPC 13. The size of a pie chart reflects the total amount of expression of fOSs. When you move the mouse cursor over the pie chart, information such as glycan structure and expression ratio are displayed for each color. Fig. 9 indicates (Hex)1(HexNAc)1 is expressed the most in WT. Knockout of the NPC1 gene increases glycan expression but markedly decreases (Hex)1(HexNAc)1 expression ratio. Administration of HPBCD, a model therapeutic drug, decreases total fOS expression to close to that of WT but notably fails to restore (Hex)1(HexNAc)1 expression although the ratio is increased. Systems such as this database for easily visualizing results will be useful for detecting changes in expression.
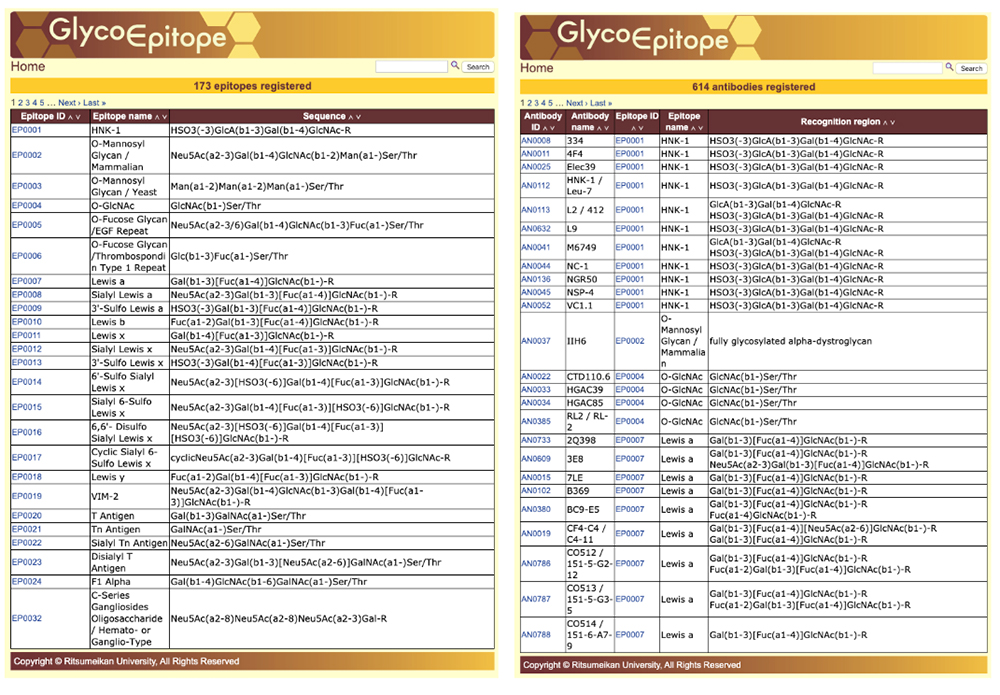
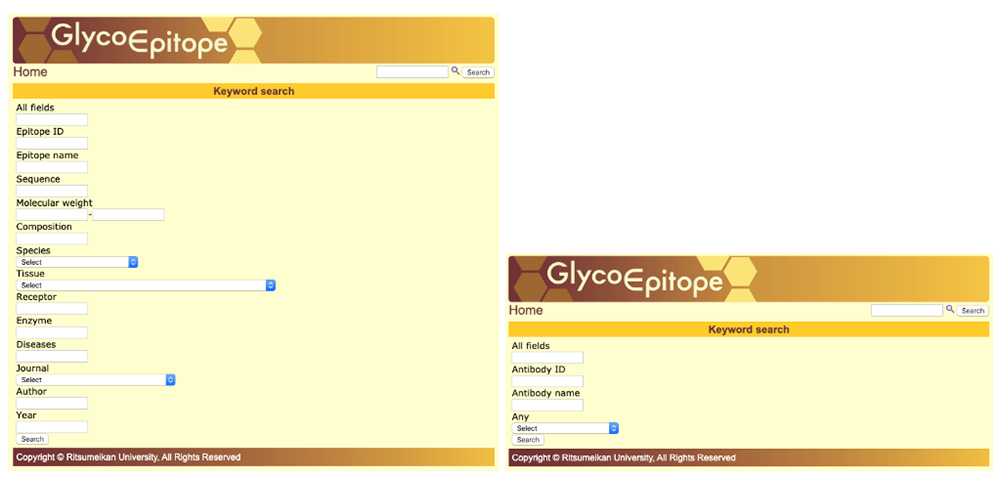
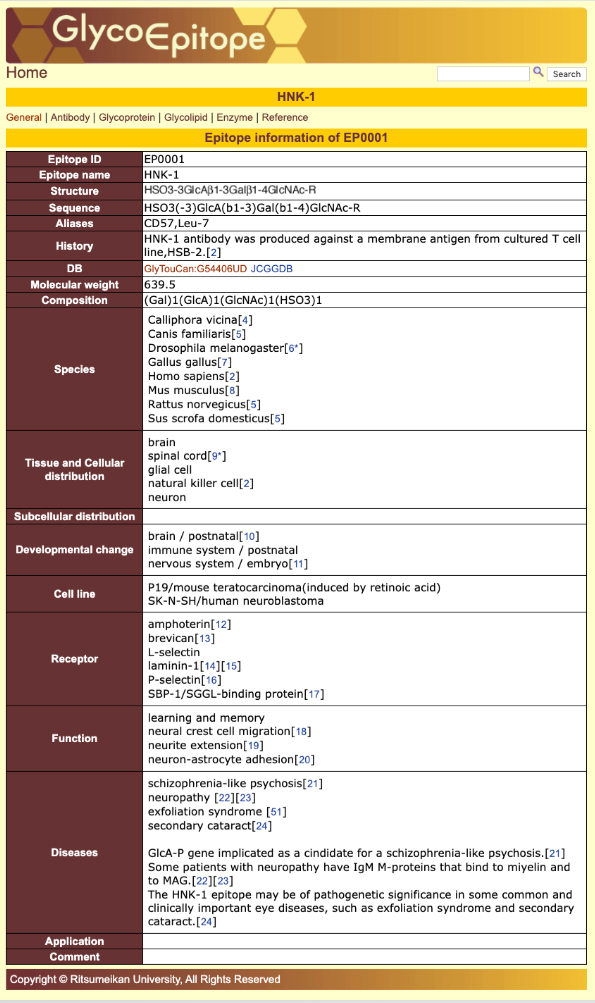
GlycoEpitope 14,15,16 is a database of information on glycan antigens and glycan-reactive antibodies. It was established with the cooperation of biologists researching glycans. This database is available from both GlyCosmos as well as https://glycoepitope.jp. GlycoEpitope contains general data on epitopes (General), antibodies that recognize epitopes (Antibody), expressed proteins (Glycoprotein), glycolipids having part of an epitope’s structure, enzymes involved in the synthesis and decomposition of epitopes, food glycans, and literature information. Information on 173 epitopes and 614 antibodies has currently been accumulated (as of October 30, 2019). Targeted information can be searched from a list of epitopes and antibodies (Fig. 10) or using a keyword search of each item (Fig. 11). The page showing the epitope details allows access to various information (Fig. 12).