
Takashi Muramatsu
Dr. Muramatsu graduated from the University of Tokyo in 1963. He was a postdoctoral fellow in the Department of Microbiology and Immunology, Albert Einstein College of Medicine, an assistant professor in the Department of Biochemistry, Kobe University School of Medicine, a visiting scientist in the Department of Molecular Biology, Pasteur Institute, and a professor in the Department of Biochemistry, Kagoshima University Faculty of Medicine. He is currently a professor in the Department of Biochemistry, Nagoya University Graduate School of Medicine. He also serves as president of the Japanese Biochemical Society and as an executive editor of Glycobiology.
Chondroitin sulfates usually contain one sulfate group per disaccharride unit. However, there are oversulfated chondroitin sulfates (Fig. 1). The first one discovered was chondroitin sulfate D in shark cartilage1,2. Its basic structure is chondroitin-6 sulfate, and many of its uronic acid residues are 2-sulfated. Later, chondroitin sulfate E was found in squid cartilage2,3. Its basic structure is chondrotin-4 sulfate, and the N-acetylgalactosamine residue frequently has a 6-sulfate group. Subsequent studies have revealed the presence of these oversulfated structures also in mammalian cells4,5 . Here the oversulfated structure found in chondroitin sulfate D will be called the D unit, and that found in chondroitin sulfate E, the E unit. The biological meaning of the oversulfated structures remained largely unknown for long time. However, recent studies have revealed that the oversulfated structures are involved in cell-surface recognition by binding to midkine6-8, chemokine9 and L-selectin9, and are implicated in the control of neurite outgrowth10. This review deals with the strong interaction of the chondroitin sulfate E structure with midkine, and its general implications. First, a brief explanation of midkine is given.

Fig. 1 Structure of Chondroitin Surfates
Midkine is a 13-kDa protein that enhances growth, migration and survival of various cells. Midkine has about 50% sequence identity with pleiotrophin / HB-GAM, but is not related to other growth factors or chemokines11,12.
Midkine is essentially composed of two domains tightly held by disulfide bridges, and each domain has three antiparallel β-sheets13. Two clusters of basic amino acids are present in the more C-terminally located domain, and serve as binding sites for glycosaminoglycans13.
Midkine expression is strong in midgestation embryogenesis, while it is detected only in restricted regions in the adult tissue12,14. However, strong expression of midkine is found in many human carcinomas and also upon inflammation and repair12, 15-17. Midkine antisense oligo DNA inhibits growth of colorectal carcinoma cells in nude mice18. Therefore, midkine is considered to contribute to the malignant phenotype of tumor cells, probably by enhancing their growth, survival and invasion. Midkine also promotes migration of inflammatory cells. In mice deficient in the midkine gene, neointima formation upon vascular injurity16 and nephritis after ischemic injury17 are significantly suppressed. Decreased migration of inflammatory cells in the deficient mice is considered to be the reason for these phenomena. On the other hand, survival-promoting activity of midkine is promising in prevention of neuronal degeneration. Retinal degeneration caused by exposure to constant light is prevented by midkine19. Delayed neuronal death of hippocampal neurons after ischemic injury is also inhibited by midkine20. Furthermore, survival of in vitro developed preimplantation embryos is enhaced by midkine21. Therefore, midkine is an important factor both as a target of molecular therapy and as a potential drug to treat degenerative diseases.
Midkine receptor is a molecular complex composed of both transmembrane proteins and transmembrene proteoglycans. A transmembrane protein, low density lipoprotein, receptor-related protein (LRP) plays important roles22. In addition, ALK tyrosine kinese has recently been reported to participate in midkine signaling23. As proteoglycans, both syndecans and a chondroitin sulfate proteoglycan are implicated to play a role in midkine signaling6,12. The intracellular signaling system of midkine includes PI3 kinase and ERK12, 24.
The interaction of chondroitin sulfate E with midkine was revealed by two independent approaches. In one side, midkine was found to bind strongly to a chondroitin sulfate proteoglycan, receptor-type protein tyrosine phosphatese ζ (PTP ζ )6, (Fig. 2). The following evidence indicates that PTP ζ is a receptor of midkine in midkine-dependent migration of embryonic neurons: 1) digestion of neurons with chondroitinase ABC eliminates the midkine action; 2) antibody to the extracellular portion of PTP ζ inhibits midkine action; 3) PTP ζ binding activity of midkine mutants correlates well with that of migration-promoting activity. PTP ζ is also involved in midkine-dependent migration of osteoblast-like cells25 and midkine-dependent survival of embryonic neurons26.
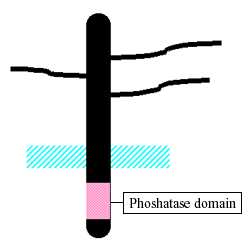
Fig. 2 Receptor-type protein tyrosine phosphatase ζ (PTP ζ )
Midkine binds to the chondroitin sulfate portion of PTP ζ with high affinity and to the protein portion with low affinity6. To discover what kind of chondroitin sulfate midkine binds strongly, inhibitory effects of various chondroitin sulfates to midkine-dependent migration were examined. Chondroitin sulfate E was found to inhibit cell migration most strongly among the various chondroitin sulfates investigated.
We also found that midkine binds to a pericellular chondroitin sulfate proteoglycan, PG-M / versican with high affinity27. The binding is inhibited by heparin, chondroitin sulfates D and E. In the chonroitin sulfate from PG-M, small amounts of D and E units have been detected.
The other line of work was started when chondroicitin sulfate E was found to inhibit neurite outgrowth on dishes coated with midkine7. Other chondroitin sulfates showed no inhibitory effects in this system28. We compared the binding affinity of chondroitin sulfate E to midkine with that of heparin to midkine, since heparin is known to bind strongly to midkine. The methods of analysis were surface plasmon resonance7 and midkine Sepharose-affinity chromatography8, in addition to inhibition of neurite outgrowth7,28. Consequently, we have reached to the conclusion that chondroitin sulfate E binds to midkine with an intensity similar to that of heparin.
We also questioned whether midkine responding cells have chondroitin sulfate rich in E units. Analysis of chondroitin sulfate synthesized by the brain from day-13 mouse embryos has revealed that a fraction of chondroitin sulfate indeed binds strongly to midkine and contains large amounts of E units8.
The other question is whether modification of chondroitin sulfate E, such as 3-O- sulfate of glucuconic acid29, is involved in strong binding to midkine. We utilized artificial chondroitin sulfate E, which was synthesized from chondroitin 4-sulfate using purified or recombinant chondroitin 4-sulfate 6-sulfotransferase30. The artificial chondroitin sulfate E was found to bind to midkine strongly, indicating that modifications such as 3-O-sulfation are not essential for strong binding8. We also noted that a cluster of E units is required for strong binding.
In case of the binding of midkine to heparin or heparan sulfate, all three sulfate groups in the heparin trisulfated unit are required for strong binding31. It is interesting that the number of sulfate residues per disaccharide unit required for strong binding to midkine is less in chondroitin sulfate E than in heparin (Fig. 3).
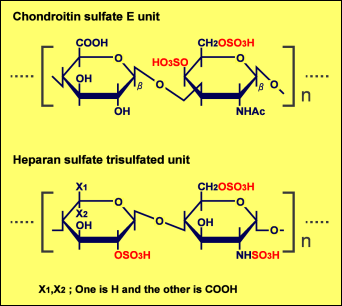
Fig. 3 Comparison of two structures with strong binding capability to midkine
Clusters of the E units are not widely distributed in the chondroitin sulfate chain. Therefore this structure can serve as a specific signal for binding to midkine as well as to other molecules.
Our studies established that clusters of chondroitin sulfate E units bind strongly to midkine and are involved in the signaling of midkine. However, the exact role of the binding in midkine signaling remains to be clarified by further studies. Most probably, midkine binding to a chondroitin sulfate proteoglycan, PTP ζ, induces dimerization of the molecule, leading to inactivation of the phosphatase activity. Since the midkine receptor is a molecular complex, the possibility that the binding to the chondroitin sulfate portion alters the relationship of the chondroitin sulfate proteoglycan to other components of the complex cannot be excluded.
The finding that chemokines9 and selectins9 also bind to chondroitin sulfate E indicates that a significant number of recognition molecules bind to oversulfated chondroitin sulfates. The classification of heparin-binding recognition molecules to those specific to heparin and heparan sulfate and those with dual recognition capability to heparin/heparan sulfate and oversulfated chondroitin sulfates is an urgent task. It has recently been reported that FGF-16, FGF-18 and HB-EGF also strongly bind to chondroitin sulfate E32.
Equally important is the establishment of the in vivo significance of recognition of oversulfated chondroitin sulfates. Chondroitin sulfates D and E are synthesizesd as outlined in Fig. 4. Indeed, knockout of chondroitin 6-sulfotransferase resulted in the absence of chondroitin sulfate D in the brain33. So far, no abnormalities have been found in the brain of the deficient mice. Most probably, the chondroitin sulfate E structure, which remained in the brain of the knockout mice compensated for the loss of the D structure.
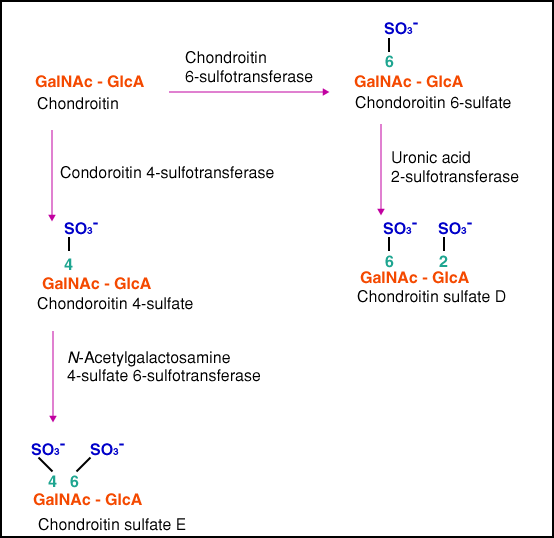
Fig. 4 Biosynthetic pathway for the formation of oversulfated chondroitin sulfates
Chondroitin sulfates:The fundamental structure of chondroitin sulfate is chondroitin, whose repeating unit is a disaccharide composed of N-acetylgalactosamine and glucuronic acid. Sulfation at different hydroxyl groups yields different chondroitin sulfates. Epimerization of the C-5 hydroxyl group in glucuronic acid of chondroitin 4-sulfate yields dermatan sulfate. Microheterogeneity in chondroitin sulfate structures is an important chracteristic. As an example, oversulfation occurs in varying degrees in chondroitin sulfates from different sources. Structural analysis of chondroitin sulfates depends on chondroitinases, a method developed by Prof. S. Suzuki. After chondoroitinase ABC digestion, chondroitin sulfates are depolymerized to unsaturated disaccharides, which are separated by HPLC and quantitated.