|
The Fbs Family: Fbs Proteins Involved in the ER-associated Degradation System
|
|
|
 |
Eukaryotic cells have two main systems for intracellular protein degradation. Autophagy is an intracellular bulk degradation process and less selective. On the other hand, ubiquitin-proteasome system accounts for most of the selective protein degradation for determination of the life span of each intracellular protein. In this ubiquitin-proteasome system, a series of enzymes, E3 ubiquitin-ligases, play the most important role in the selection of target proteins. E3s are believed to exist in hundreds of species. The largest known family of E3s is the SCF complex consisting of Skp1, Cul1, Rbx1/Roc1, and F-box proteins. In this SCF complex, only F-box proteins are variable components, and at least 100 different members of F-box proteins in human are thought to allow for the specific ubiquitylation of a wide range of substrates. In many instances, the F-box proteins serve as receptors for substrates that have various covalent modifications (phosporylation seems to be the predominant signal). Fbs1 is named as the F-box protein that recognizes sugar chains. As shown in Fig. 1, Fbs1 has an N-terminal P domain whose function is not determined, an F-box domain that is required for Skp1 binding, and a substrate-binding domain that recognizes N-glycans. |
|
|
|
|
|
Fig. 1 The SCF complex (A) and the domain structure of the Fbs family (B)
|
|
|
|
Fbs1 has the ability to bind to proteins modified with high-mannose oligosaccharides, whose modification occurs in the endoplasmic reticulum (ER). X-ray crystallographic and nuclear magnetic resonance (NMR) studies of the substrate-binding domain of Fbs1 have revealed that Fbs1 interacts with the inner chitobiose in N-glycans of glycoproteins by a small hydrophobic pocket located at the top of the 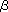 -sandwich. The intramolecular interactions of the innermost chitobiose and the polypeptide moiety generally hamper the intermolecular binding of macromolecules such as proteins. Therefore, Fbs1 might bind to denatured glycoproteins which have exposed inner chitobiose in N-glycans. Improperly folded proteins in the ER are retro-grade translocated into the cytosol where they are destroyed by the ubiquitin-proteasome system. This disposal system is called ER-association degradation (ERAD). The reason why Fbs1 interacts with internal chitobiose of N-glycans is because Fbs1 recognizes the innermost position of N-glycans as the signal for unfolded glycoproteins in the ERAD pathway ( Fig.2). |
|
|
|
|
|
|
|
Fig.2 Hypothetical model depicting the molecular mechanism of the substrate recognition of Fbs1
Although Fbs1 cannot access the inner chitobiose protected by polypeptide moieties in folded glycoproteins, Fbs1 can bind to exposed inner chitobiose by denaturation.
|
|
|
|
It has been reported that Fbs1 has at least four F-box proteins that show high homology in the substrate-binding domain. Among these proteins, Fbs1/Fgb1/Fbx2/NFB42, Fbs2/Fbg2/ Fbx6b, Fbg3/Fbx44, Fbg4/Fbx17, and Fbg5/Fbx27, it has been shown that only Fbs2 and Fbs1 have the ability to interact with N-glycans, but the N-glycan binding activities of other proteins have not been reported. Fbs2 is widely expressed in various tissues in contrast to the limited expression of Fbs1 in neural cells in adult brain (Fbs1 was first named NFB42, meaning neural F Box 42 kDa). As ERAD might be the general feature of all cells, Fbs2 is thought to be a general ERAD E3 in mammals. Although Fbs1 and Fbs2 recognize high-mannose N-glycans, Fbs1 seems to possess 102-3 higher affinity for oligosaccharides.
It has shown that both Fbs1 and Fbs2 associate with a cytosolic ATPase p97/VCP that is required for the extraction of misfolded proteins into the cytosol in the ERAD pathway. Therefore, SCF complexes that include Fbs1 or Fbs2 and p97/VCP cooperate functionally during ERAD.
|
|
|
|
Yukiko Yoshida (Tokyo Metropolitan Institute of Medical Science) |
|
|
|
|
|
| References |
(1) |
Yoshida Y: A novel role for N-glycans in the ERAD system. J. Biochem., 134, 183-190, 2003 |
|
(2) |
Mizushima T, Hirao T, Yoshida Y, Lee SJ, Chiba T, Iwai K, Yamaguchi Y, Kato K, Tsukihara T, Tanaka K: Structural basis of sugar-recognizing ubiquitin ligase. Nature Struct. Mol. Biol., 11, 365-370, 2004 |
|
(3) |
Yoshida Y, Adachi E, Fukiya K, Iwai K, Tanaka K: Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Reports, 6, 239-244, 2005 |
|
|
|
| Links |
|
GP-B07: Sweet ER Quality Control: The Role of N-glycosylation in Protein Quality Control (Sachiko Sato) |
|
|
GP-C04: N-linked Oligosaccharide as a Signal for Proteolysis (Tadashi Tai) |
|
|
ES-B01: Evolutional Cnservation on the Glycan-mediated Quality Control System for Newly Synthesized Proteins in the Endoplasmic Reticulum (ER) (Tadashi Suzuki) |
|
|
Beyond Glycogenes:  -Mannosidase-like Protein Involved in Quality control in the Endoplasmic Reticulum (ER) (Nobuko Hosokawa, Kazuhiro Nagata) -Mannosidase-like Protein Involved in Quality control in the Endoplasmic Reticulum (ER) (Nobuko Hosokawa, Kazuhiro Nagata) |
|
|
|
|
|
| Jul.5, 2006 |
|
|
|
|
|
|
|



