

 |
 |
Evolutional Conservation of the Glycan-mediated Quality Control System for Newly Synthesized Proteins in the Endoplasmic Reticulum (ER) |
||||||||||||||||||||||||||
 |
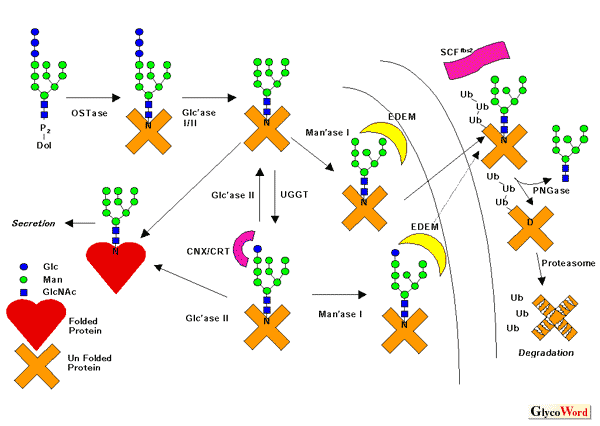
The endoplasmic reticulum (ER) is the site for biosynthesis of soluble/membrane
proteins that go through the secretory pathway. A Eukaryotic cell has
a mechanism called the "quality control system" in the ER
that distinguishes the functional proteins from aberrant ones, so that
only the former exit the compartment via vesicular transport to their
respective final destinations. Recent studies have revealed that the
asparagine(N)-linked oligosaccharides on proteins play pivotal roles
in the folding and maturation of glycoproteins. For instance, the glycan
can serve as a "tag" to represent the folding status of proteins,
so they can be aided by molecular chaperones to acquire correct folding,
or in other cases, they are targeted for degradation. The glycan-dependent
quality control machinery involves (1) transfer of the oligosaccharide
Glc3Man9GlcNAc2 from dolichylpyrophosphate
to the nascent proteins; (2) rapid deglucosylation by a-glucosidase
I/II; (3) reglucosylation of unfolded/misfolded proteins by UDP-Glucose:
glycoprotein glucosyltransferase, a folding sensor to distinguish the
folded proteins from the malfolded ones; (4) recognition of the monoglucosylated
glycans by lectins called calnexin (CNX; membrane protein) and calreticulin
(CRT; a soluble homologue of CNX) (Figure). CNX/CRT play a role as glycan-dependent
molecular chaperones, avoiding the malfolded glycoproteins to aggregate
and aiding their folding (though it is not clear at this moment if it
is a direct or indirect effect as far as folding-promoting activity
is concerned). The monoglucose recognized by these lectins is eventually
removed by The above scheme holds true for most eukaryotes, but there are a few exceptions: (a) Trypanosomatides transfer unglucosylated oligosaccharides in protein N-glycosylation, namely Man6GlcNAc2, Man7GlcNAc2 or Man9GlcNAc2, depending on the species. Therefore, the sugars on proteins are ready for glucosylation by UGGT without deglucosylation. (b) A calnexin orthologue (Cne1p) does not appear to have lectin-activity nor are there functional orthologues for UGGT in budding yeast (baker`s yeast). Accordingly there is no "calnexin cycle" in this organism. The case of budding yeast seems to be a rare exception, however. For instance, fission yeast, another yeast widely used for life science studies, has all the components mentioned above. If proteins consistently fail to achieve a correct folding state despite
the help of CNX/CRT, not only are they not functional but they can also
be toxic to cells as they form an aggregate inside the cell. Therefore,
the cell has a mechanism to eliminate such deleterious proteins by retro-translocating
them from the lumen of the ER into the cytosol, where the proteasome
degrades them. This mechanism is called ER-associated degradation (ERAD).
The EDEM protein (ER degradation enhancing |
|||||||||||||||||||||||||
 |
||||||||||||||||||||||||||
|
||||||||||||||||||||||||||
|
In mammalian cells, once the misfolded glycoproteins are exported out of the ER, glycoprotein-specific ubiquitin ligase, Fbs1/2 (Fbx2 and their paralogues) binds to the carbohydrates on the proteins and modifies them with polyubiquitin, a recognition tag for proteasomal degradation. So far no apparent orthologues of this protein family in lower eukaryotes have been reported, and it remains to be revealed how widespread the glycoprotein-specific ubiquitination is. Finally, prior to the degradation by proteasome, the N-linked glycans are removed by the action of the cytoplasmic peptide:N-glycanase (PNGase). This deglycosylating enzyme is also distributed widely in eukaryotes. However, the metabolic pathway of free oligosaccharides formed by the PNGase-action seems quite distinct between human and yeast. The molecular details of this process have not been revealed in any system. |
||||||||||||||||||||||||||
| Tadashi Suzuki (Department of Biochemistry, Graduate School of Medicine, Osaka University) | ||||||||||||||||||||||||||
|
||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||
| Dec. 28, 2004 | ||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||
|
||||||||||||||||||||||||||