|
Rapid and Accurate Identification of Oligosaccharide
Structures Using the Observational MSn Spectral Library
|
|
|
 |
Mass spectrometry (MS) is considered to be unsuitable for
the analysis of oligosaccharide structures which must distinguish a large
variety of isomers with the same molecular weight. On the other hand,
high sensitivity and high throughput of the strong points of MS, which
is an indispensable tool for proteomics, are very attractive to glycomics.
Structural analysis of oligosaccharides using MS has been achieved by
detailed assignments of the fragment ions generated from tandem mass spectrometry
(MS/MS) of the permethylated oligosaccharides. In MS/MS, ions of a particular
m/z value are selected in the first stage of mass analysis. These
"parent" or "precursor" ions are fragmented and the
product ions resulting from the fragmentation are then analyzed in the
second stage of mass analysis. Although MS/MS of permethylated oligosaccharides
affords information on linkage positions such as 1-4 or 1-3, it is impossible
to distinguish between anomers (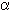 or
or 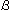 ) and diastereomers
(ex. GlcNAc or GalNAc). Additionally, permethylation requires at least
5 ) and diastereomers
(ex. GlcNAc or GalNAc). Additionally, permethylation requires at least
5 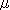 g of glycan
sample, which is equal to more than 1,000 times the protein quantity required
for protein identification in proteomics (1). g of glycan
sample, which is equal to more than 1,000 times the protein quantity required
for protein identification in proteomics (1).
However, many examples in MS/MS or multistage tandem mass spectrometry
(MSn) of native glycans (without permethylation) where different fragment
ions and/or different intensities of the same fragment ions were observed
for oligosaccharides with the same sequences, depending on the glycosidic
linkages and branching structures, have recently been reported. MSn experiments
of oligosaccharides in particular have shown that oligosaccharides may
have characteristic fragment patterns. MSn is a technique that can only
be performed on quadrupole ion-trap and FT-ICR instruments which allow
the re-fragmentation of product ions (fragment ions from MS/MS), in which
the product ions are trapped allowing another isolation and fragmentation
to be performed, resulting in the MS3 spectrum. This process can be repeated
a number of times, resulting in a series of MSn spectra where 'n' represents
the number of times the isolation-fragmentation cycle has been carried
out. Here, I introduce an example using matrix-assisted laser-desorption/ionization
quadrupole ion trap time-of-flight (MALDI-QIT-TOF) MS.
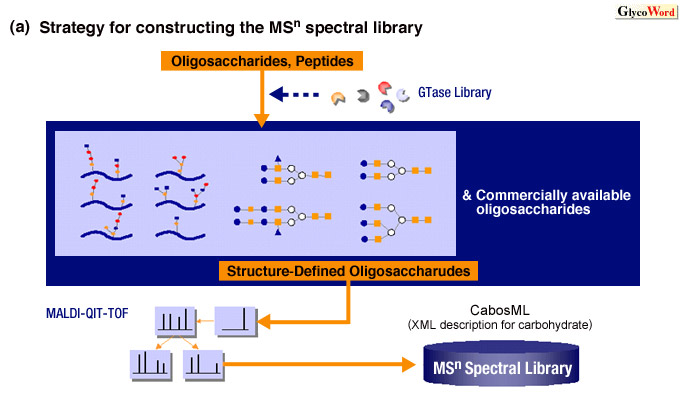
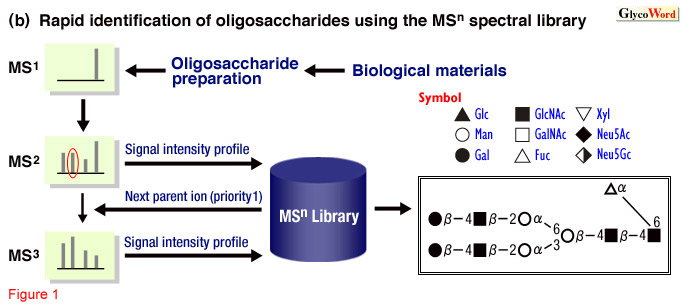
The identification of oligosaccharide structures using the observational
MSn spectral library is based on the assumption that all oligosaccharides
have distinct signal intensity profiles on their MSn spectra
(2). Rapid and accurate identification can be achieved by matching the
MSn spectra of the analyte to the observational MSn
spectral library developed by the accumulation of the MSn spectra
of a large variety (3) of structure-defined oligosaccharides (Figure
1). This strategy for oligosaccharide analysis has been realized
as shown by the recent achievements in the massive cloning of glycogenes
making possible the preparation of a variety of oligosaccharides. Accurate
identification of not only the linkage positions and branching structures
but also of anomers and diastereomers has been made possible based on
the MSn spectra acquired from 1 pmol of glycan sample. The advantage of
this strategy is that the rapid and accurate identification of oligosaccharides
can be easily performed by a researcher who is not familiar with the structural
analysis of oligosaccharides using MALDI MS, which anyone can easily operate,
without a detailed assignment of fragment ions, by consulting the MSn
spectral library. |
|
|
 |
|
 |
|
|
|
Akihiko Kameyama
(Research Center for Glycoscience, National Institute of Advanced Industrial Science and Technology (AIST) )
|
|
|
|
|
|
| References |
(1) |
Zaia J: Mass spectromtery of oligosaccharides. Mass Spectrom.
Rev, 23, 161-227, 2004 |
|
(2) |
Kameyama A, Kikuchi N, Nakaya S, Ito H, Sato T, Shikanai T, Takahashi
Y, Takahashi K, Narimatsu H: A strategy for identification of oligosaccharide
structures using observational multistage mass spectral library.
Anal. Chem, 77, 4719-4725, 2005 |
|
(3) |
Ito H, Kameyama A, Sato T, Kiyohara K, Nakahara Y, Narimatsu H:
Molecular-weight-tagged glycopeptide library: Efficient construction
and applications. Angew. Chem. Int. Ed. Engl, 44,
4547-4549, 2005 |
|
|
|
|
|
| Aug.31, 2005 |
|
|
|
|
|
|
|



