Regulation of glycosyltransferase by forming complexes | ||||||||||||||||||||||
 |
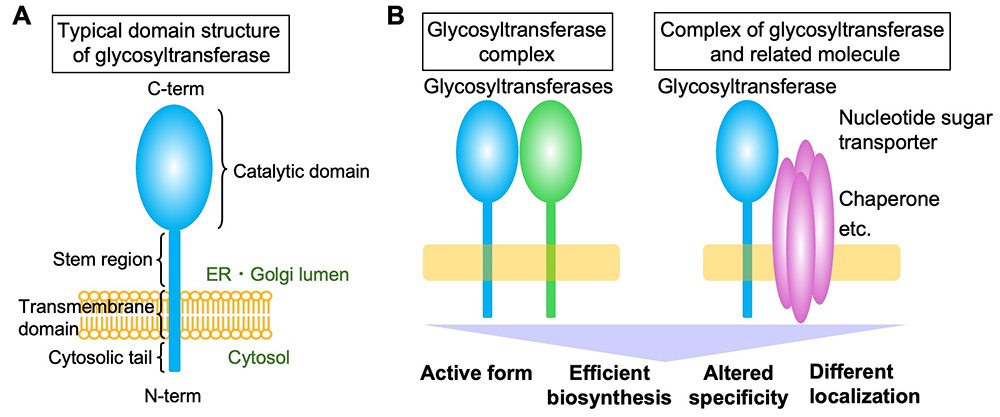
Glycan biosynthesis is catalyzed by glycosyltransferases, and mammals have approximately 200 glycosyltransferase genes. Most of these encode membrane proteins localized in the endoplasmic reticulum or Golgi apparatus, with a few exceptions such as O-GlcNAc transferase OGT. Glycosyltransferases that generate glycan precursors, glycan cores, and glycosylphosphatidylinositol (GPI) are often multi-pass membrane proteins. However, most glycosyltransferases are single-pass type 2 membrane proteins comprising a short N-terminal cytosolic tail and a large luminal C-terminal region containing a catalytic domain (Figure A). The stepwise actions of these glycosyltransferases form glycans with extremely complex structures. The Golgi apparatus is machinery that constantly generates five classes of glycans (N-glycans, O-glycans, glycosaminoglycans, GPIs, and glycolipids) with diverse structures. Glycosyltransferases, including both class-specific enzymes and enzymes common to several classes (acting on the glycan terminus in many cases), shape the glycan profiles of certain cells by sometimes competing and sometimes collaborating with each other. The glycan synthesis process does not just involve the addition of random enzyme-substrate encounters; rather, it is highly regulated by multiple factors. For instance, fine sub-Golgi localization (see the “Glycosyltransferase localization (Coming soon)” section) and recognition of specific glycoprotein substrates (see the “Selectivity of glycosyltransferases for substrate proteins” section) significantly impact glycan biosynthesis. The formation of glycosyltransferase complexes is also a crucial factor regulating glycan biosynthesis. It has been reported that glycosyltransferases form complexes with various proteins (excluding homo-oligomers comprising a single glycosyltransferase here). Several well-known examples of glycosyltransferase complexes constitute enzymes for the creation of a particular glycan (Figure B, left), thereby acquiring the activity or specificity required for synthesis of the particular glycan. For instance, the polylactosamine synthases B3GNT2 and B3GNT8 boost their activity approximately 10- and 100-fold, respectively, by forming a complex through their luminal domains (1). POMT1 and POMT2, the initial enzymes in the formation of O-Man glycans, and the heparan sulfate polymerases EXT1 and EXT2 essentially become active when they form a hetero-complex (2,3). Other enzymes for O-Man biosynthesis, POMGNT1 and FKTN, catalyze different steps in the same pathway and interact with each other via transmembrane regions (4). Similarly, the enzymes required for the multi-step reactions in the biosynthesis of human natural killer-1 (HNK-1) glycan, i.e., B4GALT2, GlcAT-P, and HNK-1ST, are reported to form complexes for efficient glycan biosynthesis (5). Based on these reports, many glycosyltransferase complexes are believed to comprise enzymes that act in one glycan biosynthetic pathway. In addition to complexes made of only glycosyltransferases, other hetero-complexes have also been reported to comprise glycosyltransferase and another relevant factors (Figure B, right). For instance, it has long been known that the substrate specificity of B4GALT1 changes upon binding to α-lactalbumin to become a lactose synthase (6). Another example is an enzyme for mucin-type glycan synthesis, C1GALT1, whose interaction with its specific dedicated chaperone, COSMC, is required for its activity (7). In addition, studies have revealed that the transporters for nucleotide sugars (the donor material for glycan synthesis) form a complex with a glycosyltransferase that utilizes the nucleotide sugars as the donor substrate. It was reported that the UDP-Gal transporter was shown to form a complex with UGT8A, the synthase for galactosylceramide, to regulate the subcellular localization of the transporter (8). More recently, an UDP-GlcNAc transporter was demonstrated to interact with a specific N-glycan-branching enzyme (9).
As these investigation on glycosyltransferase complex formation have mostly relied on overexpression in cell lines, it remains to be fully elucidated how endogenous enzymes form complexes in native tissues. Recent technological advances, such as CRISPR genome editing and proximity labeling (e.g., TurboID), have enabled tag knock-in to endogenous glycosyltransferase genes and non-IP-western blotting methods for detecting enzyme interactions. Studies using these advanced technologies to elucidate the protein complexes of endogenous glycosyltransferases in various cell types will be crucial to fully understanding the overall picture of glycan biosynthesis in cells. Yasuhiko Kizuka
|
|||||||||||||||||||||
| References | |
|---|---|
| (1) | Seko A, Yamashita K: Characterization of a novel galactose beta1,3-N-acetylglucosaminyltransferase (beta3Gn-T8): the complex formation of beta3Gn-T2 and beta3Gn-T8 enhances enzymatic activity. Glycobiology 15, 943-951, 2005 |
| (2) | McCormick C, Duncan G, Goutsos KT, Tufaro F: The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. U S A 97, 668-673, 2000 |
| (3) | Akasaka-Manya K, Manya H, Nakajima A, Kawakita M, Endo T: Physical and functional association of human protein O-mannosyltransferases 1 and 2. J. Biol. Chem. 281, 19339-19345, 2006 |
| (4) | Xiong H, Kobayashi K, Tachikawa M, Manya H, Takeda S, Chiyonobu T, Fujikake N, Wang F, Nishimoto A, Morris GE, Nagai Y, Kanagawa M, Endo T, Toda T: Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of alpha-dystroglycan. Biochem. Biophys. Res. Commun. 350, 935-941, 2006 |
| (5) | Kizuka Y, Matsui T, Takematsu H, Kozutsumi Y, Kawasaki T, Oka S: Physical and functional association of glucuronyltransferases and sulfotransferase involved in HNK-1 biosynthesis. J. Biol. Chem. 281, 13644-13651, 2006 |
| (6) | Ramakrishnan B, Qasba PK: Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the beta1,4-galactosyltransferase-I. J. Mol. Biol. 310, 205-218, 2001 |
| (7) | Ju T, Cummings RD: A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc. Natl. Acad. Sci. U S A 99, 16613-16618, 2002 |
| (8) | Sprong H, Degroote S, Nilsson T, Kawakita M, Ishida N, van der Sluijs P, van Meer G: Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell 14, 3482-3493, 2003 |
| (9) | Song W, Isaji T, Nakano M, Liang C, Fukuda T, Gu J: O-GlcNAcylation regulates beta1,4-GlcNAc-branched N-glycan biosynthesis via the OGT/SLC35A3/GnT-IV axis. FASEB J. 36, e22149, 2022 |
Mar. 15, 2024