|
Comparative Biochemistry of the Galectin Families
|
|
|
 |
Galectins comprise a family of animal lectins that bind
to  -galactoside-containing
carbohydrate moieties of glycoconjugates (See LE-A01 “Galectin:
Definition and History” in “Lectins”). Galectins have
been identified in various animal species including nematode, conger eel,
fly, zebrafish, rainbow trout, chicken, etc. In mammals, currently fourteen
galectins have been isolated and they are classified into three subgroups
according to their domain structures (galectins-1-14). We have introduced
Xenopus laevis as a model system for studying the roles of galectins,
and Xenopus is the only vertebrate whose galectin family has been
comprehensively identified, other than mammals (ref. 1, 2; Xenopus
galectin: xgalectins-Ia-VIIIa). In this issue, the author compares the
Xenopus and mammalian galectin families. -galactoside-containing
carbohydrate moieties of glycoconjugates (See LE-A01 “Galectin:
Definition and History” in “Lectins”). Galectins have
been identified in various animal species including nematode, conger eel,
fly, zebrafish, rainbow trout, chicken, etc. In mammals, currently fourteen
galectins have been isolated and they are classified into three subgroups
according to their domain structures (galectins-1-14). We have introduced
Xenopus laevis as a model system for studying the roles of galectins,
and Xenopus is the only vertebrate whose galectin family has been
comprehensively identified, other than mammals (ref. 1, 2; Xenopus
galectin: xgalectins-Ia-VIIIa). In this issue, the author compares the
Xenopus and mammalian galectin families. |
|
|
 |
|
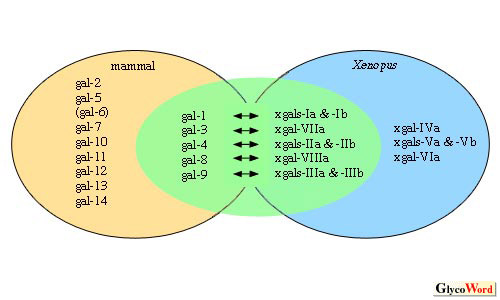
| Fig1.Members
of the Xenopus and mammalian galectin families |
Galectins in the green circle are apparent homologues
between mammals and Xenopus. Small a and b in the names of
Xenopus galectins indicate isoforms whose amino acid sequences
are highly similar.
|
|
|
|
|
|
According to the similarities of amino acid sequences
and their expression patterns, Xenopus and mammalian galectins
can be categorized into two groups in which possible homologous relationships
could or could not be determined (Fig. 1).
Xgalectin-VIIa and mammalian galectin-3 were determined to be homologous
and we have also observed similar specificity in their oligosaccharide
recognition. A representative pair with a similar expression pattern
was found when comparing the expressions of xgalectins-IIa & IIb
and galectin-4. Their mRNAs were all localized in digestive tracts.
However, the amino acid identity of every pair was limited to around
50 %. This value is lower than in the case of sequence comparison of
factors that are very essential for an organism, such as some transcription
factors, growth factors and so on. In general, we are able to obtain
identity of over 70% in the sequence comparison of these factors, whose
functions seem to be basic and universal, even between homologues of
frog and mammals.
On the other hand, there are a number of Xenopus and mammalian
galectins whose homologous relationships cannot be estimated. One representative
example is skin-galectins. In both Xenopus and mammals, there
are galectins that are specifically and abundantly expressed in skin.
These are xgalectin-Va &Vb and mammalian galectin-7. However, we cannot
estimate their homologous relationship from their amino acid sequence
similarity because the identity of sequences of xgalectin-Va and galectin-7
is only 15%. This fact reminds us that there are great differences in
the skin of Xenopus and mammals. We do not know if the difference
in skin-specific galectin is a result or a cause of the difference in
skin, but it is quite likely that the skin-specific galectins are related
to the characterization of the skin of each species. Taking into consideration,
the fact that similarities in homologous galectins between species are
relatively low, galectins seem to have not only universal functions
across species, but also seem to contribute greatly to the characterization
of each species. In fact, the galectin-7 gene is still found only in
mammals, even though DNA databases for various species have been compiled
(3). Another piece of evidence is found in the expression pattern of
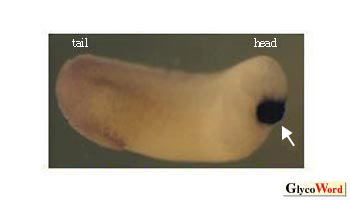
Xenopus xgalectin-VIa, whose mammalian counterpart has not been
identified. During embryogenesis, the distribution of mRNA of xgalectin-VIa
is restricted to the cement gland, which is a transient organ specific
to amphibians (Fig. 2).
|
|
|
 |
|
| Fig2.
Distribution of mRNA of xgalectin-VIa was analyzed by whole-mount
in situ hybridization. |
The mRNA was localized in the cement gland,which is indicated by an arrow.
|
|
|
The author believes that most oligosaccharide-related
genes and oligosaccharides themselves participate in the characterization
of each species. Moreover, they may also be one of the great motivating
forces behind evolution.
|
|
|
|
Hiroki Shoji (Faculty of Medicine, Kagawa University) |
|
|
|
|
|
| References |
(1) |
Shoji H, Nishi N, Hirashima M, Nakamura, T: Purification and cDNA
cloning of Xenopus liver galectins and their expression.
Glycobiology, 12, 163-172, 2002 |
|
(2) |
Shoji H, Nishi N, Hirashima M, Nakamura T: Characterization of
the Xenopus galectin family. Three structurally different types
as in mammals and regulated expression during embryogenesis. J.
Bio.l Chem., 278, 12285-12293, 2003 |
|
(3) |
Houzelstein D, Goncalves IR, Fadden AJ, Sidhu SS, Cooper DN, Drickamer
K, Leffler H, Poirier F: Phylogenetic analysis of the vertebrate
galectin family. Mol. Biol. Evol., 21, 1177-1187,
2004 |
|
|
|
|
|
|
|
|
|
|
|
|
| Jan. 24, 2005 |
|
|
|
|
|
|
|






