|
Functional Alteration of Plant AGPases
|
 |
|
 |
In the biosynthesis of bacterial glycogen and plant starch, ADPglucose as the glucose donor is formed from ATP and glucose-1-phosphate, which is catalyzed by ADPglucose pyrophosphorylase (EC 2.7.7.27; AGPase; Scheme 1).
Glucose-1-phosphate + ATP = ADPglucose + PPi (Scheme 1)
Although the bacterial and plant AGPases catalyze the same reaction, they have different structures and regulatory properties. The bacterial enzyme is composed of a single subunit, encoded by the glgC gene, that oligomerizes to form a homotetramer. In contrast, plant enzyme is a heterotetramer composed of two small and two large subunits encoded by different genes. The enzyme activity of bacterial and most plant AGPases examined is allosterically regulated. In bacteria, AGPase is allosterically activated by fructose-1,6-bisphosphate and inhibited by AMP, whereas the plant AGPase is allosteically activated by 3-phosphoglycerate (3-PGA) and inhibited by orthophosphate (Pi) (Table 1).
|
|
| Table 1. Some properties of AGPases from E. coil and higher plants |
 |
| origin |
subunit structure |
genes |
activator
|
inhibitor
|
 |
| E.coil |
homoteramer |
 |
glgC gene |
F1,6BP
|
AMP
|
| Plants |
heterotetramer |
 |
large subunit gene
small subunit gene |
3-PGA
|
Pi
|
 |
 |
|
|
|
 |
|
|
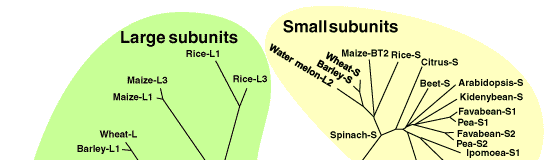
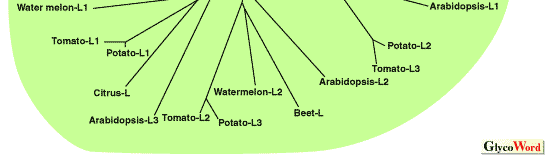
The gene sequences for large and small subunits of AGPase have been already isolated and analyzed from various plant species. Comparison of these primary seqences suggests that (1) small subunits share a very highly homology (more than 70%), (2) large subunits also have a significant homology (more than 50%) with each other, and (3) there is a relative homology (more than 40%) between small and large subunits (Fig. 1). Although the combined subunits in one enzyme have a similar molecular mass (50-60 kDa), they are distinguished as small and large subunits, because it has been demonstrated that both subunits have nonequivalent roles in enzyme function. The large subunit plays more of a regulatory role while the small subunit plays more of a catalytic role [1]. Analysis of E. coli AGPase by using chemical modifications and site-directed mutagenesis have identified the binding regions for substrates and allosteric effectors. Little has been reported on the structure-function relationships of plant AGPases with more complicated structure, but it is predicted that the substrates and allosteric molecules interact with the same regions as E. coli enzyme because these binding regions are also conserved in plant AGPases [1].
|
|
|
 |
|
 |
 |
 |
 |
Fig. 1. Phylogenetic tree of plant AGPase large and small subunits.
The alignment of the sequences was created using the CLUSTAL W program and used for constructing a phylogenetic tree. The sequences were randomly selected from the DDBJ Data Bank and expdientially named. |
|
|
 |
|
|
Evidence that the allosteric properties of plant AGPases control the quantity and quality of starch has been obtained by analyzing genetic mutant and transgenic plants. Additionally, the mutation of a large subunit gene in maize AGPase by in vivo site-specific mutagenesis led to the generation of a mutant AGPase insensitive against Pi, whereby an increase in dry seed weight of 11 to 18% was obtained [2], suggesting the possibility that the introduction of an allosterically mutant gene for AGPase into a plant may alter the seed weight or yield. A 3-D structure of plant AGPase, while still unknown, will probably lead to a theoretical alteration of the enzyme. A random mutagenesis and subsequently site-directed mutagensis approach were utilized to generate allosteric-mutants for potato tuber AGPase (Table 2) [3, 4]. For example, the mutant AGPase (UpReg-1), composed of wild type small subunit and mutated large subunit with E38K, showed an 80-fold increase in affinity for 3-PGA and a 70-fold lower affinity for the inhibitor than the wild type enzyme. Likewise, a mutant AGPase formed by introducing the same mutation into the Arabidopsis AGPase gene was insensitive to allosteric molecules. In addition, the double-mutant with both E38K and G101N might show high activity without any effectors. Details are under investigation. A transgenic plant with such an allosterically mutant gene may alter the rate for starch biosynthesis. |
|
|
|
|
|
|
 |
|
|
|
Hiroyuki Ito (Graduate School of Agriculture, Hokkaido University) |
|
|
|
|
|
| References |
(1) |
Sivak MN, Preiss J "Starch: Basic Science and Biotechnology", Adv. Food Nutr. Res. Vol. 41, 1998 |
|
(2) |
Giroux MJ et al., Proc. Natl. Acad. Sci. USA 93, 5824-5829, 1996 |
|
(3) |
Greene TW et al., Proc. Natl. Acad. Sci. USA 93, 1509-1513, 1996 |
|
(4) |
Greene TW et al., Proc. Natl. Acad. Sci. USA 95, 10322-10327, 1998 |
|
|
|
|
|
| Dec. 15, 2000 |
|
|
|
|
|
|
|



