 |
The endoplasmic reticulum (ER), where membrane and secretory proteins are newly synthesized, is the site of biosynthesis of approximately one-third of all proteins. In the ER, proteins undergo post-translational modifications such as disulfide bond formation mediated by the PDI family and N-linked glycosylation of Asn in a consensus sequence consisting of Asn-X-Ser/Thr (X≠Pro). N-glycans are attached to approximately 80% of proteins that pass through the endoplasmic reticulum. They consist of three glucoses, nine mannoses, and two N-acetylglucosamines (called G3M9) and their chains are referred to as the A, B, and C chains from left to right (see the figure). N-glycans are closely related to protein folding and degradation in the ER (see QS-A00) (1).
In higher animals, the lectin chaperones Calnexin (CNX) and Calreticulin (CRT) recognize GM9-type sugar chains, and promote proper protein conformation. Once glycoproteins with M9 attain their tertiary structure, they proceed to the secretory pathway. If proteins cannot fold properly, they are recognized by UGGT and one glucose is reattached to the M9-type N-glycan, resulting in GM9, which is a substrate for CNX and CRT to facilitate formation of the protein structure. This glucose excision and addition-dependent cycle to facilitate protein folding is called the CNX/CRT cycle (see QS-A01). This system is not highly functional in yeast.
However, proteins that cannot easily form structures must be removed because they are a burden on the endoplasmic reticulum. Their N-glycans are trimmed from M9 to M8B, and from M8B to M7, M6 and M5. When the outermost C-chain mannose is excised, unfolded or misfolded proteins are recognized by the lectin-degrading factors Yos9 in yeast and OS9/XTP3B in higher animal, are retro-translocated from the ER to the cytosol, and are degraded via the ubiquitin-proteasome system after the removal of N-glycan (1).
In yeast, trimming of mannoses from M9 to M8B in the ER is carried out promiscuously by Mns1, and trimming from M8B to M7 by the Htm1-S-S-Pdi1, a complex containing disulfide bonds. In other words, there is one rate-limiting step, and the Htm1 complex determines which proteins undergo degradation. In higher animals, four candidate molecules for mannose trimming had been mentioned: MAN1B1, EDEM1, EDEM2, and EDEM3. They share a Mannosidase Homology Domain (MHD). However, endogenous MAN1B1 was shown to localize to the Golgi (2). Analysis of endogenous gene function and enzymatic activity in vitro revealed that EDEM2 performs the trimming from M9 to M8B (3,4), and EDEM3 and EDEM1 (mainly EDEM3) perform the trimming from M8B (4,5). C558 of EDEM2 and C692 of TXNDC11 form a stable complex mediated by disulfide bonding, and the EDEM2-S-S-TXNDC11 complex, but not EDEM2 alone, exhibit strong enzymatic activity (3). TXNDC11 has five thioredoxin-like and one coiled-coil domains.
Degradation substrates will probably be selected from those with the hydrophobic side of the protein on the outside and with greater protein fluctuation. Since the substrate selection process in higher animals consists of two mannose trimming steps, from M9 to M8B and from M8B, it can be said that higher animals are more careful in selecting substrates for degradation than yeast.
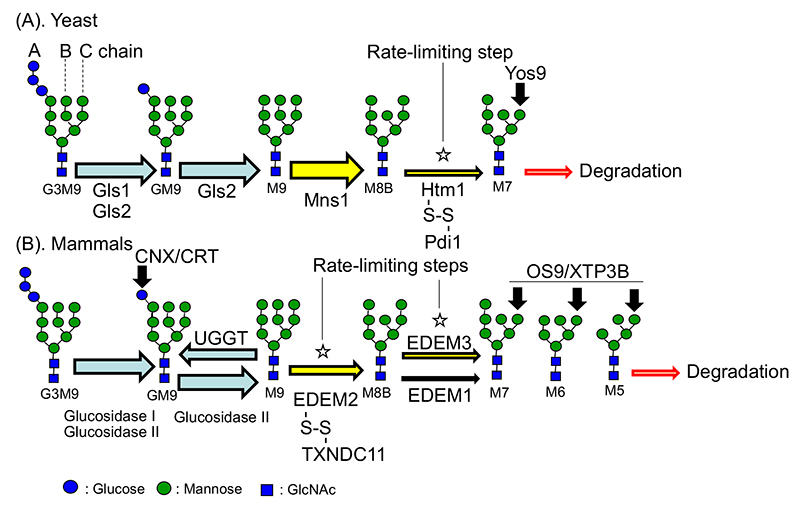
Fig. 1
(A). The chains of N-type glycans are referred to as the A, B, and C chains from left to right. In yeast, Gls1 performs glucose trimming for G3M9→G2M9 and Gls2 for G2M9→GM9→M9. From M9, Mns1 carries out mannose trimming promiscuously. Mannose trimming from M8B is carried out by the Htm1-S-S-Pdi1 complex. When the mannose in the C chain is trimmed, the trimmed-N-glycan is recognized by Yos9, which delivers the trimmed glycoprotein to degradation.
(B). In higher animals, Glucosidase I performs trimming from G3M9 to G2M9 and Glucosidase II from G2M9 to GM9 and from GM9 to M9. Once proteins attain their tertiary structure, they proceed to the secretory pathway. If they cannot form their tertiary structure, glucose is re-added by UGGT and becomes a substrate for CNX and CRT, which promote protein folding. Mannose of glycoproteins that cannot form structures is trimmed from M9 to M8B by the EDEM2-S-S-TXNDC11 complex as a first step, and from M8B to M7, M6, and M5 by EDEM3 and EDEM1 as a second step. When the mannose in the C chain is trimmed, the trimmed-N-glycan is recognized by OS9 or XTP3B, which delivers the trimmed glycoprotein to degradation. In the case of higher animals, the substrate selection process consists of two steps, mannose trimming from M9 to M8B, and from M8B to lesser extent, so it can be said that higher animals select the substrates to be degraded more carefully than yeast, in which substrates are selected by one step.
Satoshi Ninagawa
(Biosignal Research center, Kobe University)
| References |
| (1) |
Ninagawa S, George G, Mori K: Mechanisms of productive folding and endoplasmic reticulum-associated degradation of glycoproteins and non-glycoproteins. Biochim. Biophys. Acta Gen. Subj. 1865, 129812, 2021 |
| (2) |
Pan S, Wang S, Utama B, Huang L, Blok N, Estes MK, Moremen KW, Sifers RN: Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Mol. Biol. Cell 22, 2810-2822, 2011 |
| (3) |
George G, Ninagawa S, Yagi H, Saito T, Ishikawa T, Sakuma T, Yamamoto T, Imami K, Ishihama Y, Kato K, Okada T, Mori K: EDEM2 stably disulfide-bonded to TXNDC11 catalyzes the first mannose trimming step in mammalian glycoprotein ERAD. eLife 9, e53455, 2020 |
| (4) |
Ninagawa S, Okada T, Sumitomo Y, Kamiya Y, Kato K, Horimoto S, Ishikawa T, Takeda S, Sakuma T, Yamamoto T, Mori K: EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 206, 347-356, 2014 |
| (5) |
George G, Ninagawa S, Yagi H, Furukawa JI, Hashii N, Ishii-Watabe A, Deng Y, Matsushita K, Ishikawa T, Mamahit YP, Maki Y, Kajihara Y, Kato K, Okada T, Mori K: Purified EDEM3 or EDEM1 alone produces determinant oligosaccharide structures from M8B in mammalian glycoprotein ERAD. eLife 10, e70357, 2021 |
Jun. 15, 2023
|
|---|






