 |
Folding sensor function of UGGT
High-mannose-type glycans on newly generated glycoproteins in the endoplasmic reticulum are converted into diverse structures according to the folding states of the protein moiety by the action of various glycan processing enzymes. These glycans are utilized as signals to regulate “Folding acceleration”, “Folding check”, “Transport to secretion pathway” and “Transport to degradation pathway” in glycoprotein quality control. UDP-Glucose: glycoprotein glucosyltransferase (UGGT) is widely distributed in animals, plants, fungi and protozoans. This enzyme recognizes a glycoprotein having a Man9GlcNAc2-type glycan as an acceptor substrate, transfers a glucose residue at the non-reducing end of the glycan with UDP-glucose as a donor substrate, and provides Glc1Man9GlcNAc2-type glycoprotein (Fig. 1A) (1). Since UGGT also identifies the folding state of an acceptor glycoprotein and selectively glycosylates only misfolded glycoproteins, this enzyme plays an important role as a folding sensor that regulates the “Folding check” step described above. Glc1Man9GlcNAc2-type misfolded glycoproteins generated by the action of UGGT are transferred to a complex of the lectin-like molecular chaperones calnexin/calreticulin and the protein disulfide isomerase ERp57, which are responsible for the “Folding acceleration” step, and are recycled for correct folding.
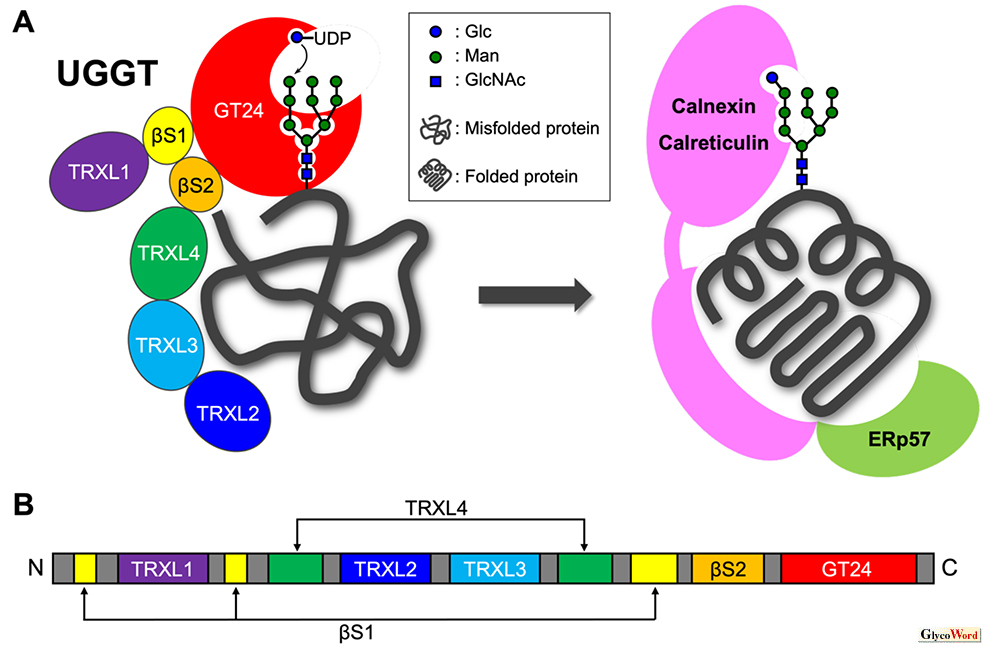
Fig. 1
(A) UGGT mediated glycoprotein glucosylation recycles glycoproteins into the complex of calnexine/calreticulin and ERp57 (B) Domain structure of UGGT
Substrate recognition of UGGT
What kind of recognition motif does UGGT require when recognizing substrate glycoproteins? Denatured natural glycoproteins such as thyroglobulin, soybean agglutinin and ribonuclease B, as well as synthetic glycoproteins that reproduce a stable misfolded state by intentionally replacing the S-S linkage, have been reported to be recognized as substrates for UGGT (2). In addition, a synthetic non-peptidic small molecule, in which a hydrophobic aglycone is introduced into Man9GlcNAc2-type glycan, has been reported to be a good substrate for UGGT (3). These findings showed that UGGT identifies a hydrophobic patch exposed on the protein surface as a recognition motif for the misfolded protein moiety. On the other hand, a recognition motif related to the glycan moiety has also been clarified. Inhibition experiments using a series of synthetic small molecules composed of different glycan moieties and a hydrophobic aglycone showed that UGGT has a strong affinity for the Man3GlcNAc2 core-structure of substrate glycoproteins having Man9GlcNAc2-type glycan (3).
Structure of UGGT
Mammalian UGGT is a soluble glycoprotein of approximately 160 kDa, composed of a catalytic domain that occupies 20% of the C-terminal side and a substrate recognition domain with a folding sensor function that occupies 80% of the N-terminal side. The catalytic domain has homology with the Glycosyltransferase family 24, and the substrate recognition domain consists of four highly flexible thioredoxin-like (TRXL) domains (Fig. 1B) (4). Two β-sandwich domains bridge the catalytic and TRXL domains. The spatial conformation between the four TRXL domains is flexible, which contributes to the accommodation of diverse misfolded glycoproteins. Two UGGT paralogs, UGGT1 and UGGT2, have been identified in vertebrates and nematodes. In the case of humans, the amino acid sequence homology between UGGT1 and UGGT2 is 55%, while the homology in the catalytic domain is over 70%. Human UGGT2 also has a transglucosylation activity similar to that of UGGT1 (5), whereas each paralog seems to be used differently depending on the target glycoprotein substrate. Furthermore, UGGT1 and UGGT2 form a 1 : 1 complex with selenoprotein F (SelenoF). Although the details of the effect of SelenoF on the function of UGGT have not been elucidated, it has been reported that SelenoF may contribute to enhancing the glycosyltransferase activity of UGGT (5).
Future perspective
UGGT has the function of selectively glucosylating Man9GlcNAc2-type misfolded glycoproteins and plays a central role in glycoprotein quality control as a folding sensor enzyme. However, the regulatory molecular mechnism for competition between UGGT and EDEMs (EDEM1, EDEM2 and EDEM3), which act on the same Man9GlcNAc2-type glycoprotein and convert the glycan into secretory signal glycan or degradation signal glycan, are still unclear. Recently, it has been shown that UGGT itself also functions as a molecular chaperone during the transglucosylation process, accelerating the folding of substrate glycoproteins (6). Further research is expected to clarify the true function of UGGT in glycoprotein quality control.
Kiichiro Totani
(Department of Science and Technology, Seikei University)
| References |
| (1) |
D’Alessio C, Caramelo JJ, Parodi AJ: UDP-Glc:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin. Cell. Dev. Biol., 21, 491-499, 2010. |
| (2) |
Izumi M, Kiuchi T, Ito Y, Kajihara Y: Misfolded glycoproteins as probes for analysis of folding sensor enzyme UDP-glucose:glycoprotein glucosyltransferase. Trends Glycosci. Glycotechnol., 25, 1-12, 2013.. |
| (3) |
Totani K, Ihara Y, Tsujimoto T, Matsuo I, Ito Y: The recognition motif of the glycoprotein-folding sensor enzyme UDP-Glc:glycoprotein glucosyltransferase. Biochemistry, 48, 2933-2940, 2009. |
| (4) |
Modenutti, CP, Capurro JIB, Ibba R, Alonzi DS, Song MN, Vasijević S, Kumar A, Chandran AV, Tax G, Marti L, Hill JC, Lia A, Hensen M, Waksman T, Rushton J, Rubichi S, Santino A, Maltí MA, Zitzmann N, Roversi P: Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: glycoprotein glucosyltransferase. Structure, 29, 357-370, 2021. |
| (5) |
Takeda Y, Seko A, Hachisu M, Daikoku S, Izumi M, Koizumi A, Fujikawa K, Kajihara Y, Ito Y: Both isoform of human UDP-glucose:glycoprotein glucosyltransferase are enzymatically active. Glycobiology, 24, 344-350, 2014. |
| (6) |
Wang N, Seko A, Takeda Y, Ito Y: Glycan dependent refolding activity of ER glucosyltransferase (UGGT). Biochem. Biophys. Acta Gen. Subj. 1864, 129709, 2020. |
Jun 15, 2023
|
|---|






