Extracellular Sulfatases SULF1 and SULF2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
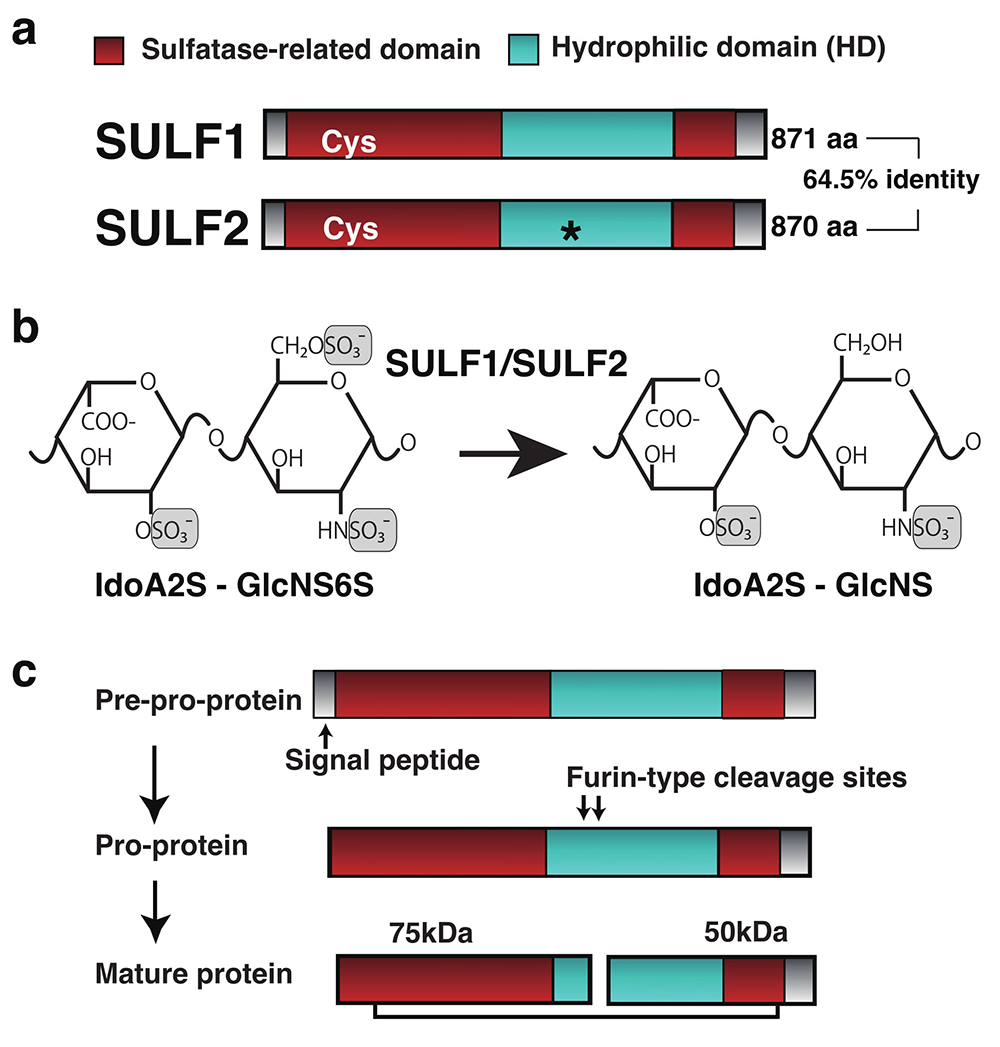
Sulfatases are enzymes that hydrolyze sulfate bonds in various molecules. Seventeen sulfatase genes have been identified in humans, and the majority of them localize to lysosomes. Lysosome-localized sulfatases sequentially degrade sulfated glycosaminoglycans and sulfated glycolipids under acidic conditions in concert with glycosidases. Human SULF1 and SULF2 were identified in 2002 as genes encoding proteins distinct from the group of intracellularly localized sulfatases (1). Human SULF1 and SULF2 proteins were shown to be secreted extracellularly and to utilize heparin/heparan sulfate (HS) chains as substrates, unlike the previously known group of intracellularly localized sulfatases (1). This is the first demonstration of extracellular endosulfatases that act on the sulfate groups within heparin/HS. SULF1 and SULF2 have been shown to release the sulfate group at position 6 of the IdoA2S-GlcNS6S unit in the highly sulfated domain called S-domain (IdoA, GlcNS, 2S, and 6S represent iduronic acid, N-sulfated glucosamine, 2-O-sulfate, and 6-O-sulfate, respectively) (Fig. 1) (1, 2). The neutral pH optimum for enzyme activity strongly supported that these enzymes work extracellularly (1).
It has been shown that SULF1 and SULF2 modulates the binding of many protein ligands to heparin or HS glycans (4, 11). In addition, SULF2 releases these ligand molecules, e.g. Vascular endothelial growth factor (VEGF) 165, C-X-C motif chemokine ligand 12 (CXCL12), C-C motif chemokine ligand 21 (CCL21), bound to immobilized-heparin from the binding complex. A mechanism has been proposed by which SULF1 or SULF2 release ligand molecules sequestered or stored in HS proteoglycans on the cell surface or in the extracellular matrix to facilitate their action on cells expressing their receptors. When Noggin, a bone morphogenetic protein (BMP) antagonist, is localized on the cell surface by binding to HS chains, BMP is tightly trapped by Noggin on the cell surface. SULF1-mediated disruption of Noggin localization at the cell surface is thought to increase in BMP bioavailability to its receptor, resulting in activation of BMP signaling. In contrast to ligand molecules that are positively regulated by SULF1/SULF2, Heparin-binding epidermal growth factor–like growth factor (HB-EGF), Fibroblast growth factor 2 (FGF2), and Hepatocyte growth factor (HGF) signaling are negatively regulated in SULF1/SULF2 expressing cells. Difficulty in formation of ligand-HS-receptor binding complex at the cell surface may induce a decrease in proliferative signaling into the cell (11-15). Phenotypic abnormalities are more severe in certain tissues during development in mice that are double deficient in Sulf1 and Sulf2 than in mice that are single deficient; the overlapping distribution of Sulf1 and Sulf2 and the ability of each Sulf to compensate for the deletion of the other has been demonstrated in mouse development (16-22). The individual functions and functional coordination of the two SULFs in different contexts, such as disease onset and progression, are topics for further study.
Kenji UCHIMURA
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |
|---|---|
| (1) | Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD: Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 277, 49175-49185, 2002 |
| (2) | Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA: Compositional profiling of heparin/heparan sulfate using mass spectrometry: Assay for specificity of a novel extracellular human endosulfatase. Glycobiology 15, 818-826, 2005 |
| (3) | Tang R, Rosen SD: Functional consequences of the subdomain organization of the sulfs. J. Biol. Chem. 284, 21505-21514, 2009 |
| (4) | Rosen SD, Lemjabbar-Alaoui H: Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin. Ther. Targets 14, 935-949, 2010 |
| (5) | Nagamine S, Keino-Masu K, Shiomi K, Masu M: Proteolytic cleavage of the rat heparan sulfate 6-O-endosulfatase SulfFP2 by furin-type proprotein convertases. Biochem. Biophys. Res. Commun. 391, 107-112, 2010 |
| (6) | El Masri R, Seffouh A, Roelants C, Seffouh I, Gout E, Perard J, Dalonneau F, Nishitsuji K, Noborn F, Nikpour M, Larson G, Cretinon Y, Friedel-Arboleas M, Uchimura K, Daniel R, Lortat-Jacob H, Filhol O, Vives RR: Extracellular endosulfatase Sulf-2 harbors a chondroitin/dermatan sulfate chain that modulates its enzyme activity. Cell Rep. 38, 110516, 2022 |
| (7) | Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP, Jr.: Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 281, 4969-4976, 2006 |
| (8) | Frese MA, Milz F, Dick M, Lamanna WC, Dierks T: Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J. Biol. Chem. 284, 28033-28044, 2009 |
| (9) | Hossain MM, Hosono-Fukao T, Tang R, Sugaya N, van Kuppevelt TH, Jenniskens GJ, Kimata K, Rosen SD, Uchimura K: Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology 20, 175-186, 2010 |
| (10) | Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP, Jr.: Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293, 1663-1666, 2001 |
| (11) | Hammond E, Khurana A, Shridhar V, Dredge K: The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front. Oncol. 4, 195, 2014 |
| (12) | Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S: Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 279, 5604-5611, 2004 |
| (13) | Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, Wong CH, Lotz MK: Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc. Natl. Acad. Sci. U S A 107, 10202-10207, 2010 |
| (14) | Wang S, Ai X, Freeman SD, Pownall ME, Lu Q, Kessler DS, Emerson CP, Jr.: QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. U S A 101, 4833-4838, 2004 | (15) | Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR: Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47, 1211-1222, 2008 |
| (16) | Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP, Jr.: SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development 134, 3327-3338, 2007 |
| (17) | Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, Carano RA, Frantz GD, Tessier-Lavigne M, Bolon B, French DM, Ashkenazi A: Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE 2, e575, 2007 |
| (18) | Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A: Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev. Dyn. 237, 339-353, 2008 |
| (19) | Hayano S, Kurosaka H, Yanagita T, Kalus I, Milz F, Ishihara Y, Islam MN, Kawanabe N, Saito M, Kamioka H, Adachi T, Dierks T, Yamashiro T: Roles of heparan sulfate sulfation in dentinogenesis. J. Biol. Chem. 287, 12217-12229, 2012 |
| (20) | Freeman SD, Keino-Masu K, Masu M, Ladher RK: Expression of the heparan sulfate 6-O-endosulfatases, Sulf1 and Sulf2, in the avian and mammalian inner ear suggests a role for sulfation during inner ear development. Dev. Dyn. 244, 168-180, 2015 |
| (21) | Takashima Y, Keino-Masu K, Yashiro H, Hara S, Suzuki T, van Kuppevelt TH, Masu M, Nagata M: Heparan sulfate 6-O-endosulfatases, Sulf1 and Sulf2, regulate glomerular integrity by modulating growth factor signaling. Am. J. Physiol. Renal Physiol. 310, F395-408, 2016 |
| (22) | Okada T, Keino-Masu K, Nagamine S, Kametani F, Ohto T, Hasegawa M, van Kuppevelt TH, Kunita S, Takahashi S, Masu M: Desulfation of Heparan Sulfate by Sulf1 and Sulf2 Is Required for Corticospinal Tract Formation. Sci. Rep. 7, 13847, 2017 | (23) | Lemjabbar-Alaoui H, van Zante A, Singer MS, Xue Q, Wang YQ, Tsay D, He B, Jablons DM, Rosen SD: Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene 29, 635-646, 2010 |
| (24) | Phillips JJ, Huillard E, Robinson AE, Ward A, Lum DH, Polley MY, Rosen SD, Rowitch DH, Werb Z: Heparan sulfate sulfatase SULF2 regulates PDGFRalpha signaling and growth in human and mouse malignant glioma. J. Clin. Invest. 122, 911-922, 2012 |
| (25) | Gill RM, Mehra V, Milford E, Dhoot GK: Short SULF1/SULF2 splice variants predominate in mammary tumours with a potential to facilitate receptor tyrosine kinase-mediated cell signalling. Cell Biol. 146, 431-444, 2016 |
| (26) | Yang Y, Ahn J, Edwards NJ, Benicky J, Rozeboom AM, Davidson B, Karamboulas C, Nixon KCJ, Ailles L, Goldman R: Extracellular Heparan 6-O-Endosulfatases SULF1 and SULF2 in Head and Neck Squamous Cell Carcinoma and Other Malignancies. Cancers (Basel) 14, 2022 |
| (27) | Luo X, Campbell NA, He L, O'Brien DR, Singer MS, Lemjabbar-Alaoui H, Ahn KS, Smoot R, Torbenson MS, Rosen SD, Roberts LR: Sulfatase 2 (SULF2) Monoclonal Antibody 5D5 Suppresses Human Cholangiocarcinoma Xenograft Growth Through Regulation of a SULF2-Platelet-Derived Growth Factor Receptor Beta-Yes-Associated Protein Signaling Axis. Hepatology 74, 1411-1428, 2021 |
Jun. 15, 2023