 |
Cellular degradation of glycosaminoglycans mainly occurs in lysosomes. Glycosaminoglycans or proteoglycans are internalized by endocytosis, and cleaved by endo-type hydrolases in endosomes or early lysosomes, to transform a very long polymer into shorter glycosaminoglycan chains or release glycosaminoglycan side chains from a core protein of proteoglycans. Glycosaminoglycan chains are further depolymerized into fragments by endo-type enzymes, and then stepwise degradation of glycosaminoglycan oligosaccharides occurs to yield monosaccharide units one-by-one, starting from the nonreducing ends by the concerted action of exo-type hydrolases within lysosomes. Inherited defects in these enzymes cause mucopolysaccharidoses (see ”Mucopolysaccharidosis”). The degradation process of each glycosaminoglycan is as follows:
1. Hyaluronan
The molecular weight (Mw) of hyaluronan is very high, at 106 – 107. After internalization into cells, hyaluronan is depolymerized into shorter molecules with Mw of 105 in endosomes by hyaluronidase 2, which is considered to be an endo-β-N-acetylglucosaminidase, and then cleaved to oligosaccharides with Mw of 104 in lysosomes by hyaluronidase 1, an endo-β-N-acetylhexosaminidase. The resultant fragments are completely hydrolyzed from the nonreducing ends to release monosaccharides by exo-type enzymes, β-glucuronidase and β-N-acetylhexosaminidase, acting in an alternating fashion (Figure 1) (1). In addition to hyaluronidases, TMEM2 as well as CEMIP/KIAA1199 have been identified as endo-type hyaluronan hydrolases. Accumulating evidence based on recent studies has suggested that they play more important roles in metabolism of hyaluronan than hyaluronidases 1 and 2 (2, 3).
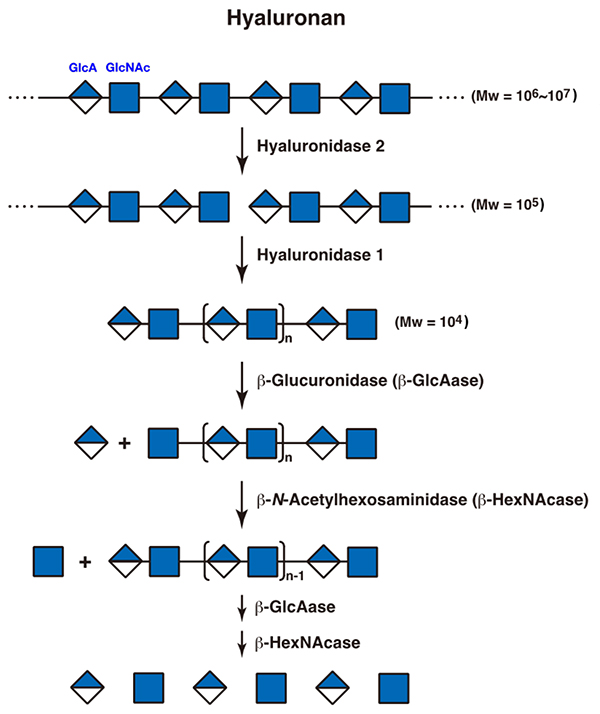
Figure 1. A scheme for the degradation of hyaluronan
GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine
|
2. Chondroitin sulfate
Chondroitin sulfate chains are also cleaved by hyaluronidase 1 to oligosaccharides with Mw of 104 within lysosomes. Since the sugar backbone of chondroitin sulfate is similar to that of hyaluronan, hyaluronidase 1 can recognize chondroitin sulfate as a substrate. The resultant oligosaccharides are completely degraded to monosaccharide units from the nonreducing ends by the concerted action of a group of exo-type hydrolases. The sequence of degradation by the exo-type enzymes acting on the oligosaccharides depends on the structure of each oligosaccharide (4, 5). Since the nonreducing terminal sugar residue of the fragments generated by cleavage of hyaluronidase 1 is nonsulfated glucuronate, exo-type β-glucuronidase acts on it first. Then, the newly formed nonreducing terminal sugar residue is recognized by an exo-type hydrolase, β-N-acetylhexosaminidase, to produce N-acetylgalactosamine. If it is sulfated, exo-type sulfatases function before the glycosidase does. When the nonreducing terminal N-acetylgalactosamine residue is 4-O- or 6-O-sulfated, N-acetylgalactosamine-4-sulfatase or 6-sulfatase hydrolyzes the sulfate group, respectively. If both positions are sulfated, N-acetylgalactosamine-6-sulfatase and then 4-sulfatase act in this order (5). Next, the exo-type β-N-acetylhexosaminidase hydrolyzes the glycosidic bond to release the N-acetylgalactosamine residue at the nonreducing end. These steps are repeated thereafter (Figure 2). Sulfate groups are generally removed before glycosidases work during these steps. However, β-N-acetylhexosaminidase can also recognize the 4-O-sulfated N-acetylgalactosamine residues (5). When the C2 position of glucuronate is sulfated, glucuronate-2-sulfatase hydrolyzes it and then β-glucuronidase cleaves the nonreducing terminal glucuronate residue. Among hyaluronidase family members, hyaluronidase 4 has been characterized as a unique hydrolase which specifically acts only on chondroitin sulfate and not on hyaluronan (6). The role of hyaluronidase 4 in the catabolism of chondroitin sulfate has not been clarified. Endo-type sulfatases isolated from some marine bacteria have been reported to hydrolyze sulfate groups from chondroitin sulfate polymers (7). Similar sulfatases have not been found in mammals.
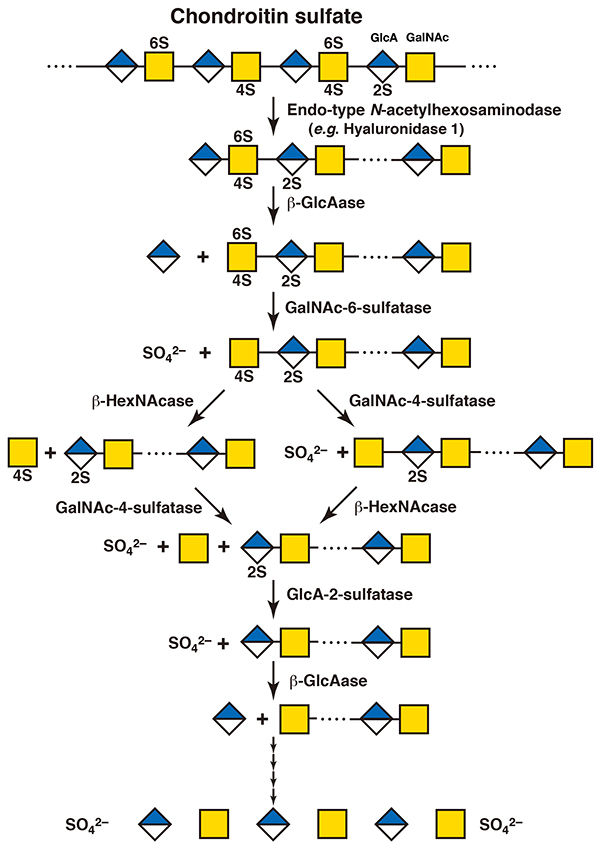
Figure 2. A scheme for the degradation of chondroitin sulfate
GlcA, glucuronic acid; GalNAc, N-acetylgalactosamine; 2S, 2-O-sulfate; 4S, 4-O-sulfate; 6S, 6-O-sulfate
|
3. Dermatan sulfate
An endo-type enzyme which specifically degrades dermatan sulfate has never been isolated. Hyaluronidases do not act on dermatan sulfate, because they recognize only the hexosaminidic bonds adjacent to glucuronic acid residues as the substrate. It remains unclear whether long polysaccharide chains of dermatan sulfate can be degraded only by exo-type enzymes from their nonreducing ends without cleavage by endo-type enzymes. Neither endo-α-iduronidase nor endo-β-N-acetylhexosaminidase which can hydrolyze the hexosaminidic bonds adjacent to iduronic acid residue, has been discovered. Dermatan sulfate oligosaccharides are degraded to monosaccharide units in the same way as chondroitin sulfate oligosaccharides (Figure 3) (4, 5). When iduronic acid is present at the nonreducing end, exo-type α-iduronidase acts on it. If it is 2-O-sulfated, iduronate-2-sulfatase works prior to α-iduronidase.
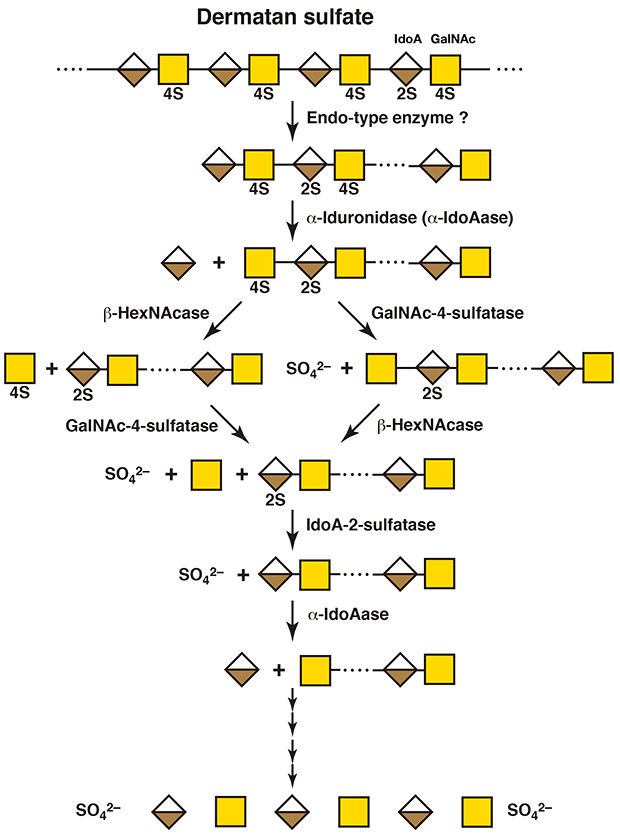
Figure 3. A scheme for the degradation of dermatan sulfate
IdoA, iduronic acid; GalNAc, N-acetylgalactosamine; 2S, 2-O-sulfate; 4S, 4-O-sulfate
|
4. Heparin/heparan sulfate
Depolymerization of heparin/heparan sulfate chains is initiated by the action of an endo-β-glucuronidase, heparanase, in endosomes or early lysosomes. Then, the resultant fragments are hydrolyzed to monosaccharide units starting from the nonreducing ends by the concerted action of exo-type enzymes within lysosomes. The sequence of degradation of the exo-type enzymes acting on them depends on their modified structure (Figure 4) (8). Since the nonreducing terminal sugar residues of the fragments generated by digestion with heparanase are sulfated glucosamines, exo-type sulfatases act on them before the exo-type glycosidase functions. If the hydroxy group at the C3 position of the glucosamine residue is sulfated, glucosamine-3-sulfatase hydrolyzes the 3-O-sulfate group. Based on the nature of the substrate specificity of heparanase, this nonreducing glucosamine residue is always 2-N-sulfated. Then, this sulfate group is hydrolyzed by the action of sulfamidase, converting the nonreducing terminal sugar residue into an N-unsubstituted glucosamine. The nonreducing terminal N-unsubstituted glucosamine residue is not hydrolyzed but N-acetylated to form an N-acetylglucosamine by transfer of an acetyl group from acetyl-CoA by the catalysis of acetyl-CoA: α-glucosaminide-N-acetyltransferase. If the hydroxy group at the C6 position of this nonreducing terminal sugar residue is sulfated, the N-acetylglucosamine-6-sulfatase cleaves this sulfate group. At length, the N-acetylglucosamine residue is released by hydrolysis with an exo-type α-N-acetylglucosainidase. Next, the new nonreducing terminal uronic acid residue is hydrolyzed by α-iduronidase or β-glucuronidase after the removal of a 2-O-sulfate group by iduronate-2-sulfatase or glucuronate-2-sulfatase, respectively, if it is 2-O-sulfated (8,9). The α-iduronidase and iduronate-2-sulfatase as well as β-glucuronidase and glucuronate-2-sulfatase are the same enzymes that act on dermatan sulfate as well as chondroitin sulfate/hyaluronan and chondroitin sulfate, respectively. Generally, the substrate specificity of exo-type enzymes is less specific than that of endo-type enzymes, and exo-type enzymes recognize only the structure of the nonreducing terminal monosaccharide moieties. Endo-type sulfatases that cleave sulfate groups from heparan sulfate/heparin polymers have been characterized (10). These are designated as heparan sulfate 6-O-endosulfatase-1 (Sulf-1) and -2 (Sulf-2), which hydrolyze a 6-O-sulfate group of 2-N-,6-O-disulfated glucosamine residues within the highly sulfated domains of the heparan sulfate side chain of proteoglycans on the cell surface (see “Extracellular Sulfatases SULF1 and SULF2”). The removal of the sulfate group results in changes in the steric conformation of the polysaccharide, which allow it to interact with different bioactive proteins, leading to the shift of its functional properties.
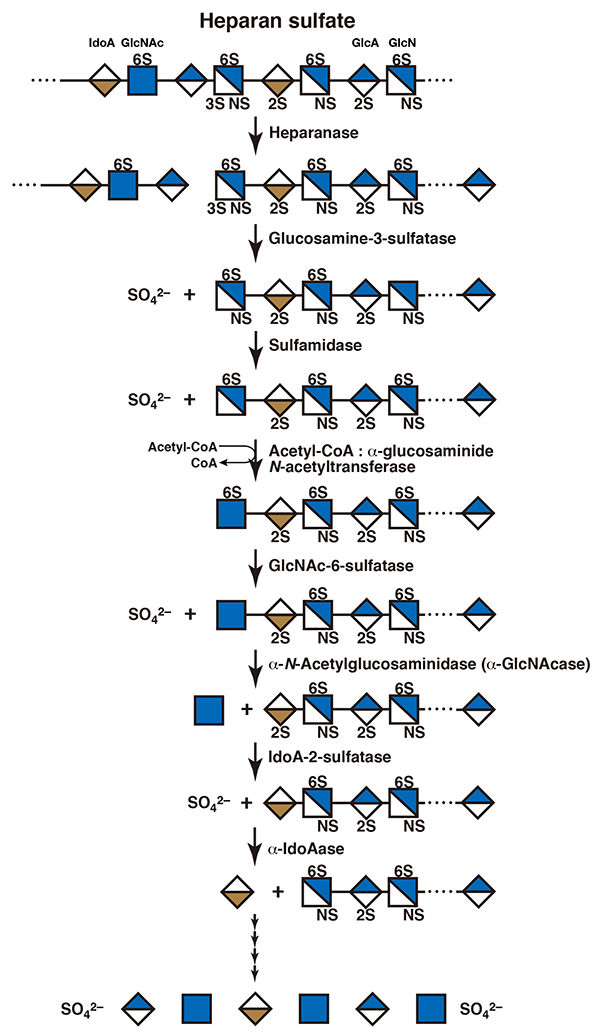
Figure 4. A scheme for the degradation of heparin/heparan sulfate
GlcA, glucuronic acid; IdoA, iduronic acid; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; NS, 2-N-sulfate; 2S, 2-O-sulfate; 3S, 3-O-sulfate; 6S, 6-O-sulfate
|
5. Keratan sulfate
Degradation of keratan sulfate also most likely initiates with depolymerization by endo-type hydrolases. An endo-β-galactosidase produces oligosaccharides. Then, the fragments are sequentially degraded from their nonreducing terminal N-acetylglucosamine residues. Exo-type enzymes, β-N-acetylhexosaminidase and β-galactosidase, act to yield monosaccharide units (11). When the nonreducing terminal sugar residue is sulfated, the sulfate groups are hydrolyzed before these glycosidases function. N-Acetylglucosamine-6-sulfatase acts on the sulfate group of the 6-O-sulfated N-acetylglucosamine residue, as described above in the section of “heparin/heparan sulfate” (12). Galactose-6-O-sulfate is recognized by galactose-6-sulfatase, which is identical to the N-acetylgalactosamine-6-sulfatase involved in the catabolism of chondroitin sulfate (Figure 5) (13).
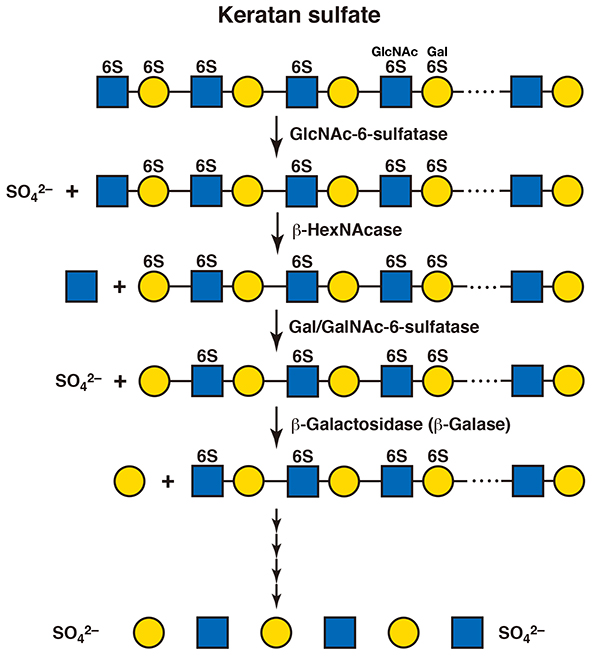
Figure 5. A scheme for the degradation of keratan sulfate
Gal, galactose; GlcNAc, N-acetylglucosamine; 6S, 6-O-sulfate
|
Shuhei Yamada
(Faculty of Pharmacy, Meijo University)
| References |
| (1) |
Kobayashi T, Chanmee T, Itano N: Hyaluronan: metabolism and function. Biomolecules 10, 1525, 2010 |
| (2) |
Yamaguchi Y, Yamamoto H, Tobisawa Y, Irie F: TMEM2: a missing link in hyaluronan catabolism identified? Matrix Biol. 78-79, 139-146, 2019 |
| (3) |
Yoshida H, Okada Y: Role of HYBID (Hyaluronan Binding Protein Involved in Hyaluronan Depolymerization), alias KIAA1199/CEMIP, in hyaluronan degradation in normal and photoaged skin. Int. J. Mol. Sci. 20, 5804, 2019 |
| (4) |
Yamada S.: Catabolism of chondroitin sulfate. Cell. Mol. Biol. Lett. 20, 196-212, 2015 |
| (5) |
Prabhakar V, Sasisekharan R: The biosynthesis and catabolism of galactosaminoglycans. Adv. Pharmacol. 53, 69-115, 2006 |
| (6) |
Yamada S, Mizumoto S: Characterization of hyaluronidase 4 involved in the catabolism of chondroitin sulfate. Molecules 27, 6103, 2022 |
| (7) |
Wang W, Shi L, Qin Y, Li F: Research and application of chondroitin sulfate/dermatan sulfate-degrading enzymes. Front. Cell Dev. Biol. 8, 560442, 2020 |
| (8) |
Hopwood JJ: Enzymes that degrade heparin and heparan sulphate. Lane DA, Lindahl U (Eds.), Heparin - Chemical and biological properties, clinical applications, Edward Arnold, London, pp. 191-227, 1989 |
| (9) |
Benetó N, Vilageliu L, Grinberg D, Canals I: Sanfilippo syndrome: molecular basis, disease models and therapeutic approaches. Int. J. Mol. Sci. 21, 7819, 2020 |
| (10) |
Vivès RR, Seffouh A, Lortat-Jacob H: Post-synthetic regulation of HS structure: The Yin and Yang of the Sulfs in cancer. Front. Oncol. 3, 331, 2013 |
| (11) |
Kresse H: Degradation of keratan sulphate proteoglycans. Greiling H, Scott JE (Eds.), Keratan sulphate – Chemistry, biology, chemical pathology, The Biochemical Society, London, pp. 111-121, 1989 |
| (12) |
Hopwood JJ, Elliott H: N-acetylglucosamine 6-sulfate residues in keratan sulfate and heparan sulfate are desulfated by the same enzyme. Biochem. Int. 6, 141-148, 1983 |
| (13) |
Glossl J, Kresse H: Impaired degradation of keratan sulphate by Morquio A fibroblasts. Biochem. J. 203, 335-338, 1982 |
Jun. 15, 2023
|
|---|











