The linkage region oligosaccharides from glycosaminoglycans
were isolated from
Drosophila melanogaster after chondroitinase
digestion and were then derivatized with 2-aminobenzamide (4). The linkage
tetrasaccharide from chondroitin sulfate was a uniform structure of
–GlcA-Gal-Gal-Xyl(2-
O-phosphate)-. In contrast, the unmodified
and phosphorylated forms were demonstrated in heparan sulfate of adult
flies at a molar ratio of 73: 27.
Three genes were recently demonstrated to affect signaling by members
of the Wnt, TGF-
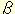
,
Hedgehog and fibroblast growth factor families in
Drosophila
encode proteins with homology to vertebrate enzymes involved in the
glycosaminoglycan synthesis (1). Detailed analysis of glycosaminoglycans
from flies bearing mutations in those three genes was carried out by
a method using enzymatic digestion and following HPLC separation (5).
It was found that mutations in
sugarless, which encodes a protein
with homology to UDP-glucose dehydrogenase, compromise the synthesis
of both chondroitin and heparan sulfate, as would be predicted from
a defect in UDP-glucronate production. Defects in
sulfateless,
a gene encoding a protein with similarity to vertebrate
N-deacetylase/
N-sulfotransferases,
did not affect chondroitin sulfate levels or composition but altered
the composition of unsaturated disaccharides from heparan sulfate.
N-,
6-
O- and 2-
O-sulfated disaccharide units were absent
and replaced entirely with an unsulfated disaccharide. A mutation in
tout-velu, a gene related to the vertebrate Exostoses 1 heparan
sulfate co-polymerase (B08 of this series), likewise did not affect
chondroitin sulfate synthesis but reduced all forms of heparan sulfate
to below the limit of detection. These findings demonstrate the utility
of
Drosophila as a model organism for studying the function
of glycosaminoglycans
in vivo.



