

 |
 |
Roles of Chitinases and beta-1,3-Glucanases in Plant Defense |
|||||||||||||||||
 |
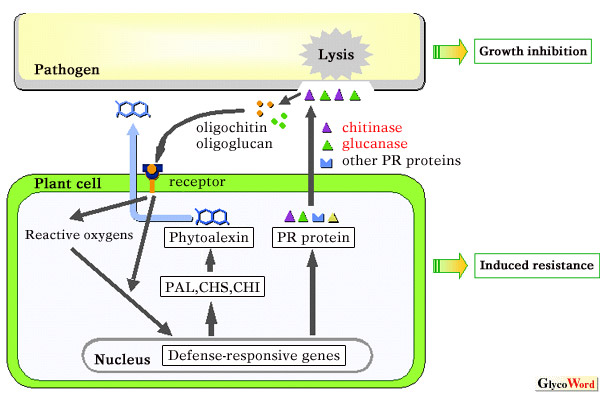
Plants activate their self-defense systems upon sensing
invading microorganisms. The induced resistance is classified into three
types: non-host resistance, variety-specific resistance mediated by R
( A part of the cell walls of various fungi including plant pathogens consists of chitin and beta-1,3-, 1-6-glucan. Many chitinases (EC 3. 2. 1. 14) and beta-1,3-glucanases (EC 3. 2. 1. 39) produced by plants can inhibit fungal growth probably by dissolving the tips of germ tubes and hyphae. It is known that a combination of chitinase and beta-1,3-glucanase shows stronger anti-fungal activity to a wider range of fungi than when each of them acts separately (1). Although there is no direct evidence to show how much plant chitinase and beta-1,3-glucanase contribute to the resistance at the infection site because they are encoded by a multigene family, a great deal of previous research has provided corroborative evidence that both enzymes play important roles in plant self-defense mechanism, i. e., plants produce several kinds of chitinase and beta-1,3-glucanase yet possess no polymer chitin, some have anti-fungal activity, expression of their genes is altered upon infection, transgenic plants overexpressing chitinase or both chitinase and beta-1,3-glucanase become less susceptible to fungal or oomycete diseases, they generate oligosaccharides, which plants recognize as so-called PAMPs and result in activation of the defense system (2) leading to the induction of resistance against fungal diseases (see “Oligosaccharide Elicitors as Crop Protectants”). Moreover, it has been reported recently that one race-specific elicitor AVR4 of the tomato pathogen Cladosporium fulvum, which is a chitin-binding protein, can protect fungi against plant chitinases (3). On the other hand, tomato evolved to recognize the AVR4 through the R gene product Cf-4 and induces race-specific (or variety-specific) resistance, which completely shuts out the fungal spread. Correspondingly, C. fulvum evaded the resistance further by mutating into AVR4, which is no longer recognized by Cf-4 but still keeps the chitin-binding activity. The interaction between tomato and C. fulvum is a very interesting example that plants have established more sophisticated systems, variety-specific resistance (or gene-for-gene resistance), by the coevolution of host (resistance) and pathogen (pathogenicity) from the defense mechanism mediated by carbohydrate recognition, which seems to be obtained prior to the evolution to plant and animal since it is conserved in both kingdoms. An other report indicates that plant chitinase genes have evolved more rapidly especially around the catalytic site than other genes, suggesting the hypothesis that plant chitinases may coevolve with fungi in response to variation in pathogen defense against chitinolytic activity (4). Future studies will provide more evidence that plants produce chitinases and beta-1,3-glucanses for self-defense and show how pathogens struggle against them. |
||||||||||||||||
|
|||||||||||||||||
| Yoko Nishizawa (National Institute of Agrobiological Sciences) | |||||||||||||||||
|
|||||||||||||||||
| Jul. 31, 2005 | |||||||||||||||||
|
|
|||||||||||||||||
|
|||||||||||||||||