 |
Determination of glycan structures is an important step in understanding the role of glycan function. Mass spectrometry (MS) currently plays a significant role in the analysis of glycan structures based on its high detection sensitivity. However, mass spectrometry is not well suited for discrimination between glycan isomers of the same molecular weight. Glycan isomerism is caused by differences in anomericity (α/β), linkage pattern (1-2/1-3/1-4/1-6, etc.), and composition (Glc/Gal/Man and GlcNAc/GalNAc, etc.). Therefore, MS-based analysis usually gives the most probable structure(s) to each mass peak based on fragment spectral data.
Separation of glycan isomers is performed mainly by liquid chromatography (LC). Various separation modes in LC have been reported for glycans, such as normal-phase, anion-exchange and reversed-phase. Multidimensional separations combining these modes have also been performed. Among them, hydrophilic interaction liquid chromatography (HILIC), which uses a highly polar stationary phase, is widely used to separate a wide mass range of samples. Recently, ion mobility spectrometry (IMS) has been applied as a complementary approach to separating glycan isomers (1). IMS can be regarded as electrophoresis in the gas phase. The sample is ionized and transferred into a drift tube partially filled with helium or nitrogen gas under the influence of an electric field. The ionized sample is separated based on its collision cross section (CCS, Ω), charge number (z), and mass (Figure 1). Drift velocity (Vd) is proportional to the drift voltage (E) and has the following relation to ion mobility (K).
Vd = K×E
This equation can also be expressed using charge number (z) and collision cross section (Ω).
Vd = C (z/Ω)E (C is a constant)
Here, Ω is the CCS between the ionized sample and the buffer gas, which is unique to the ion under the same gas pressure. Therefore, in principle, glycan isomers of equal mass can be separated provided that their CCSs are different (Figure 2).
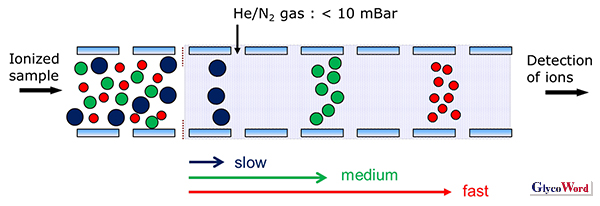
Figure 1
Principle of ion mobility spectrometry
|
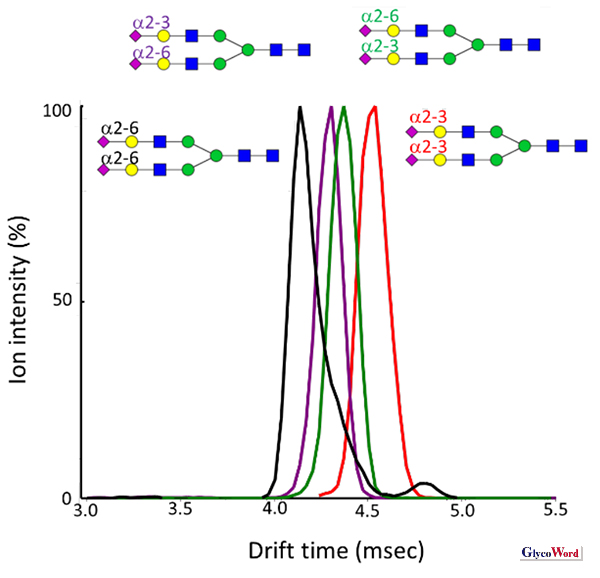
Figure 2
Separation of sialoglycan isomers by IMS
|
CCS can be calculated from the time to travel the drift tube (drift time, tD). Since CCS is correlated with the 3D structure of glycans, experimental and theoretical studies are being carried out to investigate 3D structure-CCS relationships. A weak correlation has been reported between the degree of N-glycan branching and CCS (2). Molecular dynamics simulations of several branched N-linked glycans have also been performed and their CCSs were calculated. The predicted CCS values are in good agreement with experimentally measured CCS values (3). If the CCS of each glycan is predicted computationally, it will greatly help in the IMS analysis of unknown glycans. Conversely, 3D structural information on glycans will be obtained from the experimentally-determined CCS. Thus, IMS is expected not only to be a means of isomer separation of glycans, but also to contribute to solving the 3D structure of glycans.
IMS can be easily combined with MS or LC/MS because IMS separation is attained on the millisecond timescale. Such combinations make it possible to correlate CCS with LC elution time and molecular mass. The combined use of these approaches is expected to enable rapid identification of glycan structures. Ion mobility mass spectrometry (IM/MS) instruments were commercially launched in 2006, and IM/MS has become a common technique. To improve the resolution of IM/MS, cyclic ion mobility (cIM) device has been developed recently, in which sample ions go into a circular path to increase their traveling distance (4). Repeated cycling theoretically increases the resolution of cIM.
To facilitate rapid and accurate identification of glycan structures, a database is necessary integrating glycan-related information. The GlycoMob database is now available which contains CCS data of glycans (5). There is also a report of dataset for N-linked glycans conjugated with the pyridylamino (PA) group as a fluorophore (Figure 3) (2). CCS, retention time and m/z data can be utilized to identify the glycans by liquid chromatography/ion mobility mass spectrometry (LC/IM/MS). Since more than 1,000 N-linked glycans are estimated to be found in proteins, glycan data needs to be collected and added to the database.
Reliable and rapid identification of glycan structures by incorporating ion mobility spectrometry will lead to a deeper understanding of the functional aspects of glycans. Furthermore, it will be useful for qualitative evaluation of biopharmaceuticals.
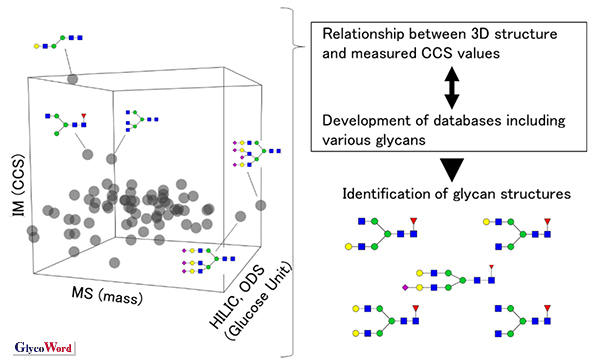
Figure 3
Concept of database development and glycan structure identification
|
Noriyoshi Manabe
(Tohoku Medical and Pharmaceutical University, Institute of Molecular Biomembrane and Glycobiology, Division of Structural Glycobiology )
| References |
| (1) |
Yamaguchi Y, Nishima W, Re S, Sugita Y: Confident identification of isomeric N-glycan structures by combined ion mobility mass spectrometry and hydrophilic interaction liquid chromatography. Rapid Commun. Mass Spectrom. 26, 2877-2884, 2012 |
| (2) |
Manabe N, Ohno S, Matsumoto K, Kawase T, Hirose K, Masuda K, Yamaguchi Y: A Data Set of Ion Mobility Collision Cross Sections and Liquid Chromatography Retention Times from 71 Pyridylaminated N-Linked Oligosaccharides. J. Am. Soc. Mass Spectrom. 33, 1772–1783, 2022 |
| (3) |
Re S, Watabe S, Nishima W, Muneyuki E, Yamaguchi Y, MacKerell AD Jr, Sugita Y: Characterization of Conformational Ensembles of Protonated N-glycans in the Gas-Phase. Sci. Rep. 8, 1644, 2018 |
(4) |
Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, Langridge D, Green M: A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 91, 8564-8573, 2019 |
(5) |
Struwe WB, Pagel K, Benesch JL, Harvey DJ, Campbell MP: GlycoMob: an ion mobility-mass spectrometry collision cross section database for glycomics. Glycoconj. J. 33, 399-404, 2016 |
Jun. 15, 2023
|
|---|









