 |
Hyaluronan (HA) is a non-sulfated glycosaminoglycan composed of repeating disaccharide units of ᴅ-glucuronic acid (GlcA) and N-acetyl-ᴅ-glucosamine (GlcNAc). HA serves as a major constituent of the extracellular matrix in vertebrates and acts as a capsular polysaccharide to evade the host immune system in certain bacteria, such as Streptococcus pyogenes and Pasteurella multocida. The enzyme responsible for the biosynthesis of this polysaccharide is hyaluronan synthase (HAS). Based on similarities in amino acid sequence and predicted structure of the proteins, HASs are classified into two families: Classes I and II. The HASs of Streptococci, Chlorella virus, and vertebrates belong to the Class I family, whereas pmHAS of Pasteurella multocida belongs to the Class II family (1, 2). Class I and II HASs differ in their structure, membrane topology, and HA biosynthetic mechanisms.
Class I HASs are glycosyltransferases classified in the same glycosyltransferase family 2 (GT2 family) as cellulose synthase (EC 2.4.1.12) and chitin synthase (EC 2.4.1.16) (http://www.cazy.org/GT2.html). Class I HAS has multiple transmembrane and membrane-bound domains composed of hydrophobic amino acids at the amino- and carboxy-terminal sides (Fig. 1). The central part of the molecule contains catalytic sites with conserved D, DxD, and QxxRW motifs. These motifs are required for the alternate transfer of monosaccharides from the sugar donors, uridine diphosphate (UDP)-GlcNAc and UDP-GlcA, to the elongating HA (Fig. 1). The three-dimensional structure of the Chlorella virus HAS (Cv-HAS) revealed by cryo-electron microscopic analysis is in good agreement with the results of a topological analysis, in which the transmembrane domains are inserted into the plasma membrane and the catalytic sites are oriented toward the cytoplasm (3). The results of the structural analysis also suggest that binding and hydrolysis of the UDP-GlcNAc substrate and GlcNAc priming cause a conformational change in the transmembrane domain of this enzyme, forming a new transmembrane channel for HA secretion (Fig. 2). Cv-HAS is monomeric and elongates HA from the GlcNAc primer toward the non-reducing end (Fig. 2), whereas bacterial HAS has been reported to dimerize and elongate the reducing end of HA (4). On the other hand, Class II HAS consists of a membrane attachment domain near the carboxy-terminus and two domains with glycosyltransferase activities that elongate the non-reducing end of the HA chain (4).
Vertebrate HASs include three isoforms, namely HAS1–3, which differ in their activity, the elongation rates of their products, and stability (5). Among the mammalian HASs, it has been reported that the activity and turnover of HAS2 are highly regulated by post-translational modifications such as phosphorylation, O-GlcNAcylation, and ubiquitination (6). Furthermore, the natural antisense transcript HAS2-AS1 regulates HAS2 mRNA stabilization (6). Thus, HA synthesis is highly regulated at multiple steps involving transcription, translation, and post-translational modifications of various HASs.
The identification of HAS genes and the recent determination of the three-dimensional structure of Cv-HAS have provided new insights into the mechanism of HA biosynthesis. However, numerous issues regarding the direction of HA chain elongation, HA secretion, and HAS regulation remain to be elucidated.
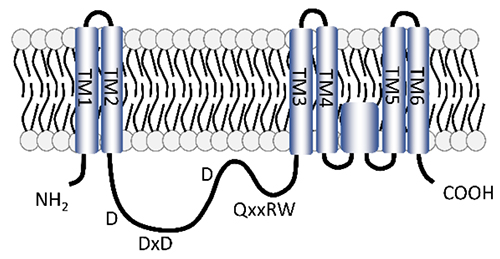
Fig. 1. Predicted structure of a vertebrate HAS
TM1-6 represent transmembrane domains, and D, x, R, and W represent aspartic acid, any amino acid, arginine, and tryptophan, respectively.
|
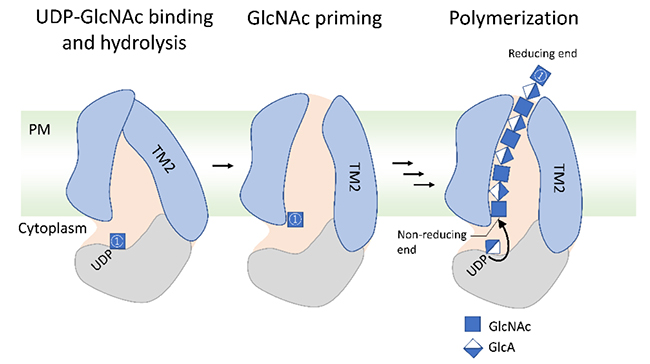
Fig. 2. Proposed model for HA elongation and secretion (Example of Cv-HAS).
Binding and hydrolysis of the UDP-GlcNAc substrate and GlcNAc priming cause a conformational change in the transmembrane domain, forming a new transmembrane channel for HA secretion. Starting from the GlcNAc primer, monosaccharides from UDP-GlcA and UDP-GlcNAc are alternately transferred to synthesize HA. The synthesized HA is thought to be secreted from the cell through the transmembrane channel. A GlcNAc primer is denoted as ①. PM: plasma membrane, TM2: transmembrane domain 2.
|
Naoki Itano
(Faculty of Life Sciences, Kyoto Sangyo University)
| References |
| (1) |
Weigel PH: Functional characteristics and catalytic mechanisms of the bacterial hyaluronan synthases. IUBMB Life 54, 201-211, 2002 |
| (2) |
Itano N, Kimata K: Mammalian hyaluronan synthases. IUBMB Life 54, 195-199, 2002 |
| (3) |
Maloney FP, Kuklewicz J, Corey RA, Bi Y, Ho R, Mateusiak L, Pardon E, Steyaert J, Stansfeld PJ, Zimmer J: Structure, substrate recognition and initiation of hyaluronan synthase. Nature 604, 195-201, 2022 |
| (4) |
Weigel PH, DeAngelis PL: Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J. Biol. Chem. 282, 36777-36781, 2007 |
| (5) |
Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, Miyauchi S, Spicer AP, McDonald JA, Kimata K: Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J. Biol. Chem. 274, 25085-25092, 1999 |
| (6) |
Vigetti D, Karousou E, Viola M, Deleonibus S, De Luca G, Passi A: Hyaluronan: biosynthesis and signaling. Biochim. Biophys. Acta 1840, 2452-2459, 2014 |
Jun. 15, 2023
|
|---|








