|
Enzymatic synthesis of glycosphingolipids and their derivatives
|
|
|
 |
Glycosphingolipids (GSLs), amphipathic compounds consisting of oligosaccharide and ceramide (Cer) moieties, have been defined as tumor antigens, receptors for microbes and their toxins, and possible modulators of various cellular activities. Recently, GSLs were found to be enriched with cholesterol and GPI-anchor proteins to form lipid microdomains called rafts on the plasma membrane of vertebrates. Sialic acid-containing GSLs, gangliosides, are relatively abundant in neural cells and considered to play significant roles in the nervous system. Arecent study indicated that knockout mice of UDP-glucose:Cer:glucosyltransferase (GlcT) gene showed embryonic lethality, suggesting that GSLs are integral molecules for early embryogenesis of mammals, although the molecular base for the GlcT-deficient lethality during early embriogenesis has yet to be clarified.
In this paper, the author describes the enzymatic synthesis of GSLs and their derivatives using two unique enzymes, endoglycoceramidase (EGCase) and sphingolipid Cer N-deacylase (SCDase) (Fig. 1). EGCase (EC 3.2.1.123, also called Cer glycanase) is a glycohydrolase which hydrolyzes the linkage between the oligosaccharide and Cer of various GSLs (Fig. 1). The enzyme was first found in actinomycetes Rhodococcus sp., then in the leech, short-necked clam and jellyfish. The genes encoding EGCase were cloned from Rhodococcus sp. M-750 and jellyfish in our laboratory. Site-directed mutagenesis and homology modeling of Rhodococcus EGCase using Clostridium A/5 cellulase as a template demonstrated that both EGCase and A/5 cellulase share a common catalytic domain in the form of a (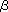 / / )8-barrel. This result indicates that EGCase catalyzes the general acid/base reactions in a similar manner for family A/5 cellulases and suggests that both enzymes are generated from the same ancestral gene. )8-barrel. This result indicates that EGCase catalyzes the general acid/base reactions in a similar manner for family A/5 cellulases and suggests that both enzymes are generated from the same ancestral gene. |
|
|
|
|
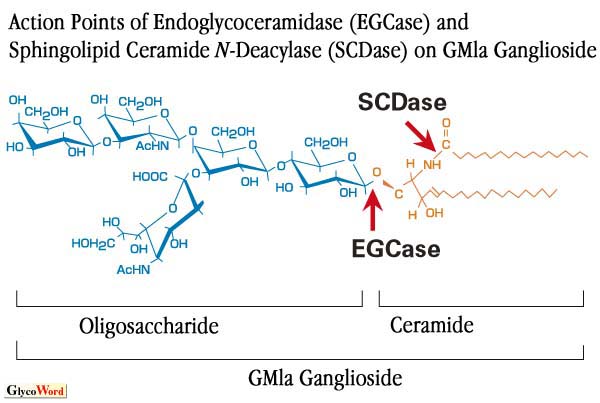
Fig. 1
|
|
|
|
|
Using the EGCase, one can easily obtain intact oligosacharides and Cers simultaneously from various GSLs. Here, the synthesis and labeling of GSLs using the transglycosylation and reverse hydrolysis (condensation) reactions of jellyfish EGCase are dscribed. Various alkyl-GM1 oligosaccharides (alkyl-II3NeuAcGgOse4) are synthesized when GM1 ganglioside is treated with EGCase in the presence of 1-alkanols. Among various 1-alkanols tested, methanol is found to be the most preferential acceptor, followed by 1-hexanol and 1-pentanol. GM1 is the best donor, followed by GD1b and GT1b, when methanol is used as an acceptor. However, neither globoside nor glucosylCer is utilized by the enzyme as a donor substrate. The enzyme transfers oligosaccharides from various GSLs to NBD-Cer, a fluorescent Cer, producing NBD-labeled GSLs. In addition to the transglycosylation reaction, the enzyme catalyzes the reverse hydrolysis reaction; lactose is condensed to Cer to generate lactosylCer in the presence of the enzyme. These results indicate that the jellyfish EGCase will facilitate the synthesis of various neoglycoconjugates and GSLs (Fig. 2). |
|
|
|
|
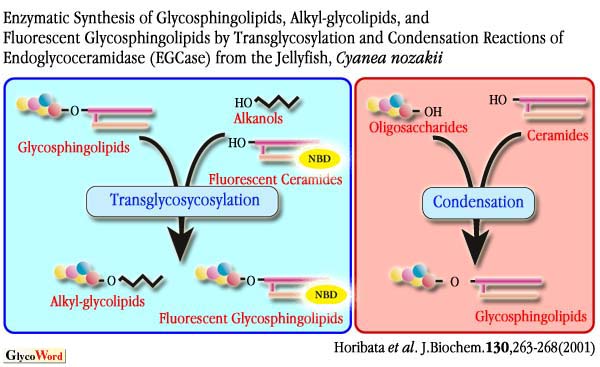
Fig. 2 |
|
|
|
|
More than 400 species of GSLs possessing different sugar structures have been reported. It should be noted, however, that GSLs show heterogeneity not only in their sugar chain but also in their Cer moieties. The biological significance of Cer heterogeneity is still not well understood. However, the structure of Cer, especially the fatty acid moieties, could influence the localization and functions of GSLs on the plasma membrane, possibly by direct interaction with cholesterol, phospholipids, and the transmembrane domains of receptor proteins. Here, the new technology using SCDase for remodeling of fatty acid moiety of GSLs is described.
SCDase is an enzyme which hydrolyzes the N-acyl linkage of the Cer moiety of various GSLs and sphingomyelin (Fig. 1).This enzyme was found in the culture supernatants of several bacteria and cloned from the genomic library of Shewanella alga. We found that SCDase efficiently catalyzes a reverse hydrolysis (condensation) reaction in which fatty acids are condensed to lyso forms of GSLs (GSLs without fatty acids). The hydrolysis of GSLs by the enzyme proceeds efficiently at acidic pH in the presence of high concentrations of detergents, whereas the reverse reaction tends to be favored at neutral pH with a decrease of the detergent concentration. Fatty acids are removed from various GSLs utilizing the hydrolysis reaction of SCDase in the presence of 1% Triton X-100 with 70-80% yield. After purification of lyso-GSLs using a reverse phase C18 and silica gel HPLC, various saturated or unsaturated fatty acids are condensed with lyso-GSLs by the reverse hydrolysis reaction of SCDase in the presence of 0.1% Triton X-100. SCDase shows a wide specificity for acceptor lyso-GSLs, i.e. all lyso-GSLs tested and lyso-sphingmyelin (sphingosylphosphocholine) are found to be good acceptor substrates for the enzyme. The reaction rate exceeds 70% for saturated fatty acids as a substrate, while it retains 50-60% for unsaturated fatty acids. Using the present method, we successfully obtained GM1a reconstructed with a single fatty acid such as palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2),  -linolenic acid (C18:3), EPA (C20:5), and DHA (C22:6) (Fig. 3). Interestingly, only lyso-sphingolipids with L-erythro-configuration, but not those with D-threo-, L-erythro and L-threo-configurations, are efficient acceptors. -linolenic acid (C18:3), EPA (C20:5), and DHA (C22:6) (Fig. 3). Interestingly, only lyso-sphingolipids with L-erythro-configuration, but not those with D-threo-, L-erythro and L-threo-configurations, are efficient acceptors. |
|
|
|
|
Fig. 3
|
|
|
|
|
14C-Labeled fatty acids are efficiently condensed to lyso-GSLs yielding 14C-GSLs in 25 mM phosphate buffer (pH 7.0) in the presence of SCDase. However, under this condition  -aminododecanoic acid is hardly condensed to lyso-GSLs by SCDase. Once the -aminododecanoic acid is hardly condensed to lyso-GSLs by SCDase. Once the  -amino group is blocked with trifluoroaetate (TFAc) and the reaction pH is made alkaline, the reaction proceeds efficiently showing an optimum pH of 10. After reaction, the N-TFAc is removed from N-TFAc-amino-GSLs by treatment with sodium methoxide. Finally, NBD-labeled GSLs are prepared from -amino group is blocked with trifluoroaetate (TFAc) and the reaction pH is made alkaline, the reaction proceeds efficiently showing an optimum pH of 10. After reaction, the N-TFAc is removed from N-TFAc-amino-GSLs by treatment with sodium methoxide. Finally, NBD-labeled GSLs are prepared from  -amino-GSLs by coupling with 4-fluoro-NBD (Fig. 3). -amino-GSLs by coupling with 4-fluoro-NBD (Fig. 3).
The present methods using EGCase and SCDase are novel, much easier, and give high yields in comparison with conventional methods. Furthermore, the fact that a wide range of sample amount from nano mol to milli mol can be used is significantly advantageous. Thus the synthesis of GSLs and their derivatives using these enzymes should be valuable for future GSL research. |
|
|
|
Makoto Ito (Department of Bioscience and Biotechnology, Kyushu University) |
|
|
|
|
|
| References |
(1) |
Ito M, Yamagata T, : A novel glycosphingolipid-degrading enzyme cleaves the linkage between the oligosaccharide and ceramide of neutral and acidic glycosphingolipids. J. Biol. Chem. 261, 14278-14282, 1986 |
|
|
(2) |
Sakaguchi K, Okino N, Izu H, Ito M, : The Glu residue in the conserved Asn-Glu-Pro sequence of endoglycoceramidase is essential for enzymatic activity. Biochem. Biophys. Res. Commun. 260, 89-93, 1999 |
|
(3) |
Horibata Y, Okino N, Ichinose S, Omori A, Ito M, : Purification, characterization and cDNA
cloning of a novel acidic endoglycoceramidase from the jellyfish, Cyanea nozakii. J. Biol. Chem. 275, 31297-31304, 2000 |
|
|
(4) |
Horibata H, Higashi H, Ito M, : Transglycosylation and reverse hydrolysis reactions of endoglycoceramidase from the Jellyfish, Cyanea nozakii. J. Biochem. 130, 263-268, 2001 |
|
(5) |
Ito M, Kurita T, Kita K, : A novel enzyme that cleaves the N-acyl linkage of ceramides in various glycosphingolipids as well as spingomyelin to produce their lyso forms. J. Biol. Chem. 270, 24370-24374, 1995 |
|
|
(6) |
Furusato M, Sueyoshi N, Mitsutake S, Sakaguchi K, Okino N, Ito M, : Molecular cloning and characterization of sphingolipid ceramide N-deacylase from a marine bacterium, Shewanella alga G8. J. Biol. Chem. 276, 17300-17307, 2002 |
|
(7) |
Kita K, Kurita T, Ito M, : Characterization of the reversible nature of the reaction catalyzed by sphingolipid ceramide N-deacylase. - A novel form of reverse hydrolysis reaction -Eur. J. Biochem. 268, 1-12, 2001 |
|
(8) |
Nakagawa T, Tani M, Kita K, Ito M, : Synthesis of fluorescent GM1 and SM utilizing the reverse hydrolysis reaction of sphingolipid ceramide N-deacylase. -Use of fluorescent substrates for assays of sphingolipid degrading enzymes and sphingolipid-binding proteins- J. Biochem. 126, 111-119, 1999 |
|
|
|
|
|
| Sep.12, 2002 |
|
|
|
|
|
|



