
Kenji Kadomatsu
Trustee, Tokai National Higher Education and Research System; Provost, Nagoya University; Director, Institute for Glyco-core Research
He graduated from Kyushu University School of Medicine in 1982 and joined the Department of Pediatric Surgery; received his M.D. in 1989, was an NIH fellow in 1990, and returned to Japan in 1993. He became a professor (Department of Biochemistry) at Nagoya University Graduate School of Medicine in 2004 and was appointed Dean of the same school in 2017; became Director of the Institute for Glyco-core Research in 2021 and Trustee, Tokai National Higher Education and Research System; Provost, Nagoya University in 2022. In recent years, his research has focused on glycans and neuronal axon regeneration. Seeking a broader perspective, he proposed the Human Glycome Atlas Project, which he continues to work on to this day.
The Human Glycome Atlas Project (HGA) started in Japan in April 2023. This project will raise the level of glycan information availability, which has been remarkably low relative to that of other major biopolymers (nucleic acids and proteins). For this purpose, human glycan structures and their biosynthesis mechanisms will be comprehensively obtained within the HGA. Human glycan structures, together with clinical information and phenotypes, will be catalogued as individual data of a large population. This information will be stored in the knowledgebase TOHSA, and all researchers worldwide will be able to easily incorporate glycan information into their routine research. Thus, the HGA will give scientists a new vision of life and open a new era of life science.
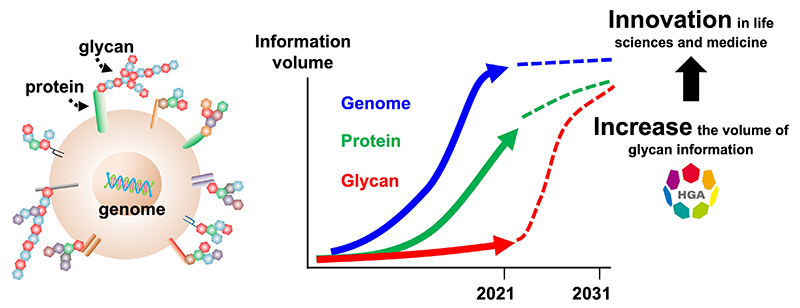
The earth is home to 8.7 million species of living organisms, equivalent to about 550 gigatons of carbon as biomass. Of this, plants account for 450 gigatons, and animals only 2 gigatons. Furthermore, humans account for 0.06 gigatons, or only 0.01% of the earth's biomass1. The molecules that make up all these life forms are remarkably similar. For example, nucleic acids (DNA and RNA: chains of nucleotides), proteins (chains of amino acids), and glycans (chains of monosaccharides) are common biomolecules across all domains of life. However, when we compare the amount of information on these three major biological chains of molecules, the amount of information on glycans is far less than that on nucleic acids and proteins. This has severely hindered our understanding of life.
The surface of every cell, without exception, is covered with a forest of glycans. When each of the 37 trillion cells that make up the human being encounters a virus or other foreign substance, or when cells encounter each other, glycans are therefore at the forefront. In fact, the anti-influenza drug Tamiflu was created to target a glycoenzyme that acts on cell surface glycans, and glycans are also essential for SARS-CoV-2 infection. Furthermore, the ABO blood type is defined by glycans, and many cancer markers are glycans or molecules with glycans. In this way, glycans contribute to all realms of life described to date. However, we know very little about glycans, because the amount of basic information of glycans remains limited. Therefore, most researchers have avoided glycans in their attempts to understand life.
The biggest bottleneck in glycoscience has been the complexity and diversity of glycan structures. However, the technological basis for solving this problem is now being established. Therefore, we have decided to launch an infrastructure project to obtain the structures of all human glycans and to raise the level of availability of glycan information to that of nucleic acids and proteins (Fig. 1). We were able to start the Human Glycome Atlas Project (HGA) in April 2023 as a Large-scale Academic Frontier Promotion Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (https://www.mext.go.jp/a_menu/kyoten/20230727-mxt_kouhou02-1.pdf)2. The Large-scale Academic Frontier Promotion Project is an infrastructure development project for academia supported by MEXT. Until now, most of these funded projects have been in the fields of physics and space science, as represented by the Kamiokande and Alma telescope projects. The HGA is the first project in the life science field.
With the HGA, we can fill in the last piece of the jigsaw puzzle of life: glycans. By being able to freely utilize glycan information, we will be able to see a new picture of life emerge. This will lead to the development of preventive and therapeutic methods for dementia, aging, intractable cancer, diabetic nephropathy, and other diseases that have been unattainable until now.
The Human Genome Project, arguably the most significant life science infrastructure project to date, began in 1990 as a U.S.-led effort to sequence the entire genome of a single human being. Five countries, the United Kingdom, Japan, France, Germany, and China, also joined this Project, sharing the task of deciphering the human genome. Celera Corporation also undertook a similar project on its own. This sparked competition, but it also gave rise to the Bermuda Principles (February 1996), which are based on the principle of data openness and the sharing of analytical techniques, consequently accelerating project completion. In 2001, both parties published draft whole-genome sequences3,4. This established the discipline and revealed the complexity of genome structure, eventually leading to the explosive expansion of molecular biology with the discovery of epigenomes, non-coding RNAs, and other new mechanisms related to the human genome. It was as if scientists had gained a new vision of life and the contribution of the Human Genome Project to life science had become immeasurable. We are now entering an era in which genome medicine is the norm, as exemplified by the SARS-CoV-2 RNA vaccine and preventive cancer treatment based on genomic diagnosis. Thus, the contribution of the Human Genome Project to society is also immeasurable.
Projects to develop infrastructure for the second life chain, proteins, has also left a significant footprint. The Protein 3000 Project, supported by MEXT, promoted three-dimensional structural analysis of proteins from 2002 to 2006 (http://www.tanpaku.org/e_index.php). The Human Protein Atlas, which began in 2000 and is still ongoing, is a Swedish initiative with India, Korea, Japan, China, Germany, France, Switzerland, the United States, Canada, Denmark, Finland, the Netherlands, Spain, and Italy participating. They have developed antibodies, advanced trans-omics, and clarified the spatio-temporal distribution of proteins to create a valuable knowledge database that is freely available to the community. Many life scientists use this database on a daily basis (https://www.proteinatlas.org/).
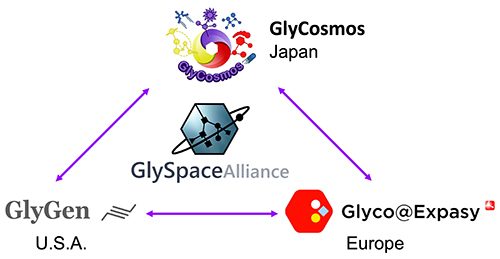
For the third life chain, glycans, the United States5, Japan6, and Europe (https://www.ludger.com/images/news/A-roadmap-for-Glycoscience-in-Europe.pdf) have created their own roadmaps to guide glycoscience research. The US, Japan, and Europe have been advancing glycan informatics independently, but in 2018 the GlySpace Alliance was started to collaborate and integrate glycoscience data with each other (http://www.glyspace.org/) (Fig. 2). Glycans, through the HGA, are about to embark on the same journey as the above-mentioned life chain infrastructure projects.
The vision of the HGA, which aims to revolutionize life science by pushing the information amount of human glycans to the level of nucleic acids and proteins, is lofty and requires appropriate preparations to realize it. Three requirements are particularly important: (1) Competence, (2) Organization, and (3) Plan.
Glycoscience, like other life science fields, has developed through international collaboration and competition. For example, about 60% of glycan-related genes have been identified by Japanese scientists, and many glycan analysis technologies originated in Japan. Even today, Japan has world-class capabilities in core technologies for the HGA, such as analysis and informatics. The HGA will eventually achieve harmonization of infrastructure development for glycans with the rest of the world, and we hope to bring about a revolution in the life sciences by involving researchers in other fields.
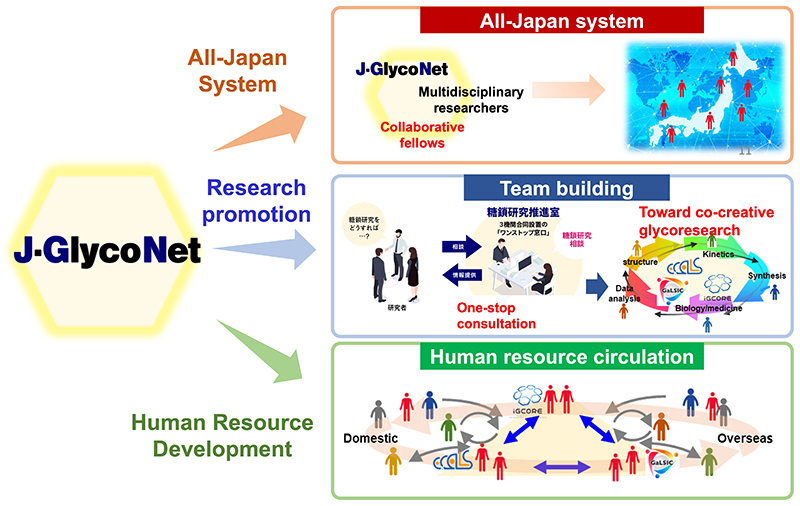
Establishment of an all-Japan system: J-GlycoNet, a joint usage/research center supported by MEXT, was launched in April 2022 (https://j-glyconet.jp/) (Fig. 3). First, J-GlycoNet provides a foundation for an all-Japan system, in which more than 100 independent glycoscientists from all over Japan work as collaborative fellows, to promote joint research with researchers in other fields. Second, it provides research support. J-GlycoNet supports research team building through one-stop consultation to increase the awareness of researchers in other fields. The third is to nurture the next generation of glycoscientists through seminars and other activities.
Gaining the support of the research community: The HGA has gained strong support from Japanese academic societies and federations of academic societies with a combined membership of 200,000 including the Japanese Biochemical Society, the Molecular Biology Society of Japan, The Japanese Society of Carbohydrate Research, Japan Consortium for Glycobiology and Glycotechnology, Polysaccharides Forum with Great Future, Japan Society for Bioscience, Biotechnology, and Agrochemistry, the Japan Neuroscience Society, the Japanese Society for Bioinformatics, the Union of Japanese Society for Biological Sciences (an association of 32 Japanese life science-related societies), and Union of Brain Science Association in Japan (an association of 31 Japanese brain science-related societies).
International networking: Collaborative agreements were established with Lille University (France), Griffith University (Australia), Macquarie University (Australia), Academia Sinica (Taiwan), Harvard University (USA), and the Genos Glycoscience Research Institute (Croatia). In addition, the GlySpace Alliance, a global glyco-informatics consortium, and CarboMet, a European glycoscience consortium, have pledged their support. The HGA has already appeared at several international conferences, such as Glyco26 (2023) in Taiwan and the 2023 Annual Meeting of the Society for Glycobiology in the U.S., where it received great anticipation and support.
Establishment of HGA research system: Tokai National Higher Education and Research System (a university corporation consisting of Nagoya University and Gifu University) is the main implementer of HGA, and the National Institutes of Natural Sciences and Soka University will also play a central role in the project (https://human-glycome-atlas.org/). Based on the all-Japan system under J-GlycoNet, the project will collaborate with cohort research institutions holding biobanks, institutions with glycan analysis technologies, and others. As an important aspect of the planning and operation of the project, we are strongly aware that the results of this project will be used not only by glycoscientists but also by researchers in a wide range of related scientific fields.
Model: The Human Genome Project not only decoded the genome sequence, but also contributed to the development of DNA sequencing technology, and as previously mentioned, had a great impact on both life science and society. The HGA is the glycan version of the Human Genome Project, and the vision and plan for this project were formulated following the Human Genome Project approach. Creation of a valuable community resource similar to The Human Protein Atlas database is the model of the knowledgebase TOHSA which will be built under the HGA. We would like to build a knowledgebase that all researchers involved in the life sciences, regardless of field, can freely utilize without hesitation.
Scientific goal: The central dogma of life is DNA→RNA→protein. On the other hand, it is impossible to understand life by the central dogma alone. Most life scientists know this. It is important to know the functions of molecular groups beyond the control of the central dogma. The most difficult part of this "extended central dogma," is the accurate incorporation of the understanding of glycans. The HGA will provide the basis for clarifying the reality of the extended central dogma, which is the scientific goal of this project.
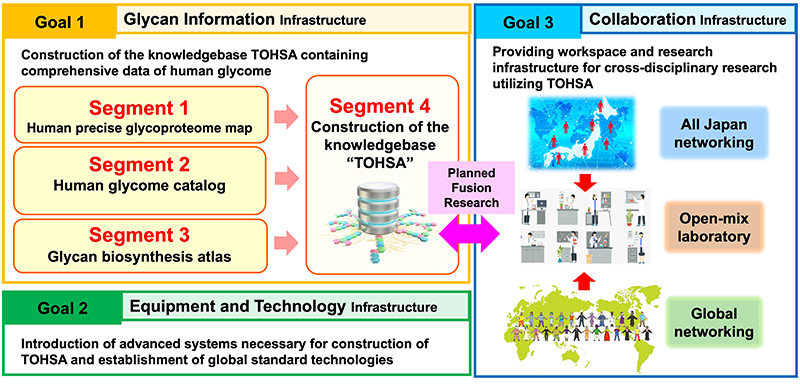
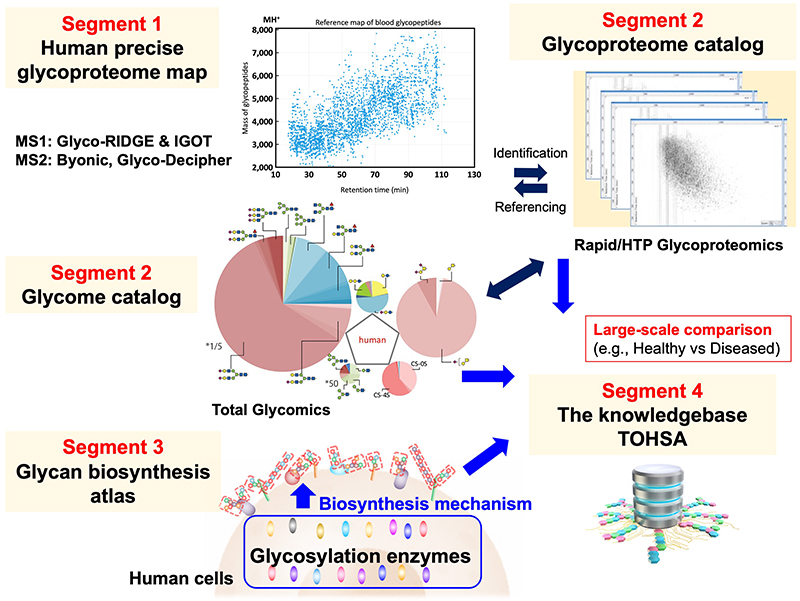
Measures to reach the scientific goal: To reach this scientific goal, we have established four segments (Figs. 4 and 5).
Segment 1. Creation of a precise reference map of human glycans. This will be done through powerful qualitative glycoproteomics, emerging analytical technologies that are able to survey the entire glycoproteome in a single experiment.
Segment 2: Creation of a human glycans catalog. We will use the reference map to perform individual quantitative glycoproteomic analyses of large populations. In addition, through a “Total Glycomics” approach, glycan structures of five categories of N-linked glycans, O-linked glycans, glycolipids, glycosaminoglycans, and free oligosaccharides can be simultaneously obtained for each individual. To obtain these glycan structure data, we will collaborate with a range of research institutes that manage biobanks. In this way, individual glycan structures can be catalogued together with clinical information and phenotypes. Since glycans vary from individual to individual and from disease to disease, this will allow us to identify the characteristics of glycans according to disease, race, age, gender, and so on.
Segment 3: Elucidation of the mechanisms of glycan biosynthesis. The activities of all glycosylation enzymes involved in human glycan biosynthesis will be standardized and mapped for their subcellular localization. This will enable prediction of glycan structures and bespoke modifications of the glycan biosynthesis pathways for medical applications.
Segment 4: The data obtained in Segments 1-3 above will be stored together with clinical data and phenotypes in the knowledgebase TOHSA, which will have user and application programming interfaces that can be easily used by all researchers and shared worldwide.
Segments 1-4 will be executed within Research Goal 1, “Establishment of Glycan Information Infrastructure”. On the other hand, “Establishment of Equipment and Technology Infrastructure” will be carried out under Research Goal 2 to support these segments. Furthermore, to achieve the highest possible impact of this project and the data resource provided by TOHSA, planned fusion research will be carried out within Research Goal 3, “Establishment of Collaborative Infrastructure”. For example, we aim to elucidate the relationship between diseases and glycans through joint research with clinical medical researchers. Similarly, trans-omics research involving glycans will be conducted in collaboration with genome researchers, and the production of glycan-modified cells will be conducted in collaboration with cell biologists and clinical medicine researchers.
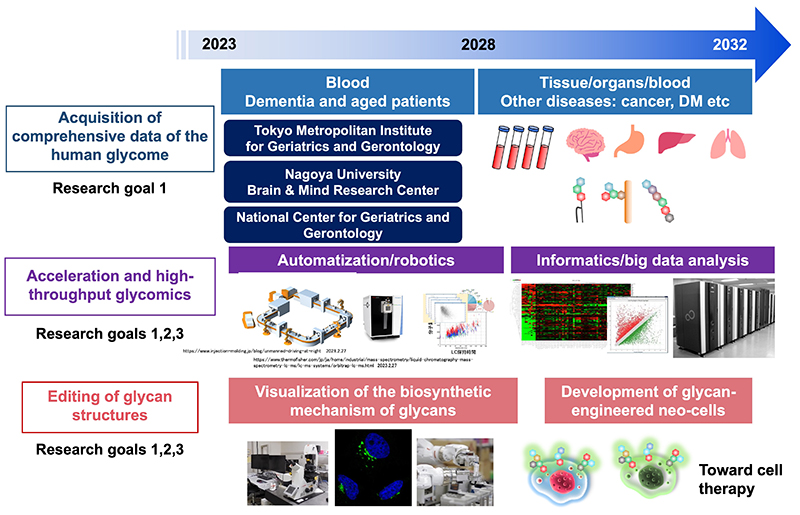
Annual Plan: During the first five years, the research will focus on dementia and aging, and 20,000 samples, mainly plasma and serum, will be analyzed (Fig. 6). During the next five years, the research target will be expanded to include intractable cancer, diabetes and other diseases, and 200,000 samples from postmortem brain, cancer tissue, normal tissue, etc. in addition to plasma and serum, will be analyzed. Automatic analyzers will be developed with the goal of analyzing a large number of samples and making analysis possible even for non-specialists. In addition, informatics technologies, which have long been considered a bottleneck for progress in the field, will be improved. Technologies for editing glycan structures will also be developed to create glycan-modified cells that can be applied to medicine.
Characteristics of glycan information in the HGA: The characteristics of glycan information in the knowledgebase TOHSA can be summarized as follows: 1. Comprehensive glycoproteomics is challenging and thus the compilation of robust and curated glycoproteome data, when completed, will be extremely useful; 2. “Total glycomics” that simultaneously obtains glycan structures across five categories is unique in the world and will lead to a comprehensive view of the entire glycan “forest” in human health and disease; 3. Comprehensive analysis of the activities and localization of glycosyltransferases is the first attempt, and will greatly improve the prediction of glycan structures based on gene expression; 4. The completion of a personalized glycans catalog with clinical information and phenotypes on as many as 200,000 cases will be unprecedented in the world, and will be useful for the classification, prediction, diagnosis, prevention, and treatment of dementia, aging, and other diseases.
The Glycoforum will regularly post information on the HGA in the future. Part of this information will include progress reports on the HGA. We make note that the HGA is not for glycoscience alone. Its purpose is to support and inspire researchers across many fields to incorporate glycan information into their own daily research and eventually obtain a nuanced, more holistic picture of life. We hope that countries around the world will participate in this project and that it will become a major movement in the life sciences.
I would like to thank Chihiro Sato, Hiromune Ando, Tetsuya Okajima, Yasuhiko Kizuka, Kiyoko Flora Aoki-Kinoshita, and Morten Thaysen-Andersen for their critical comments on this manuscript.